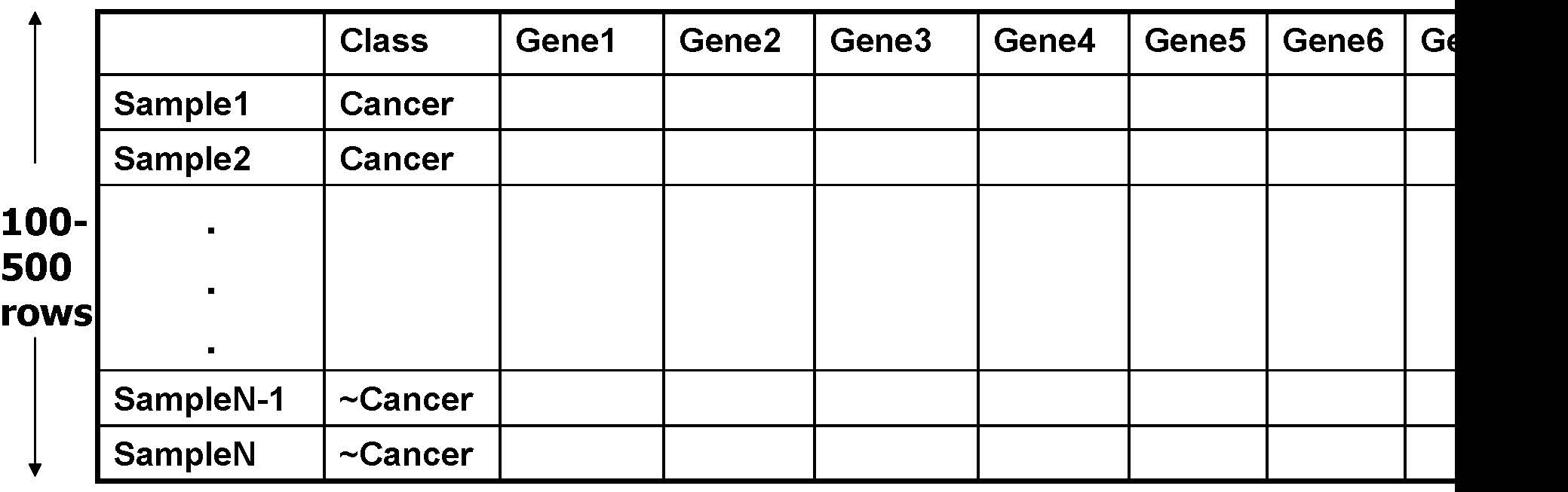
A Microarray Dataset
1000 -100,000 columns
- •Find closed patterns which occur frequently among genes.
Sample-Wise Enumeration Methods for Mining Microarray Data
Anthony K. H. Tung
Department of Computer Science
National University of Singapore
A Microarray Dataset
1000 -100,000 columns

Challenge I
“a”however not part of the group
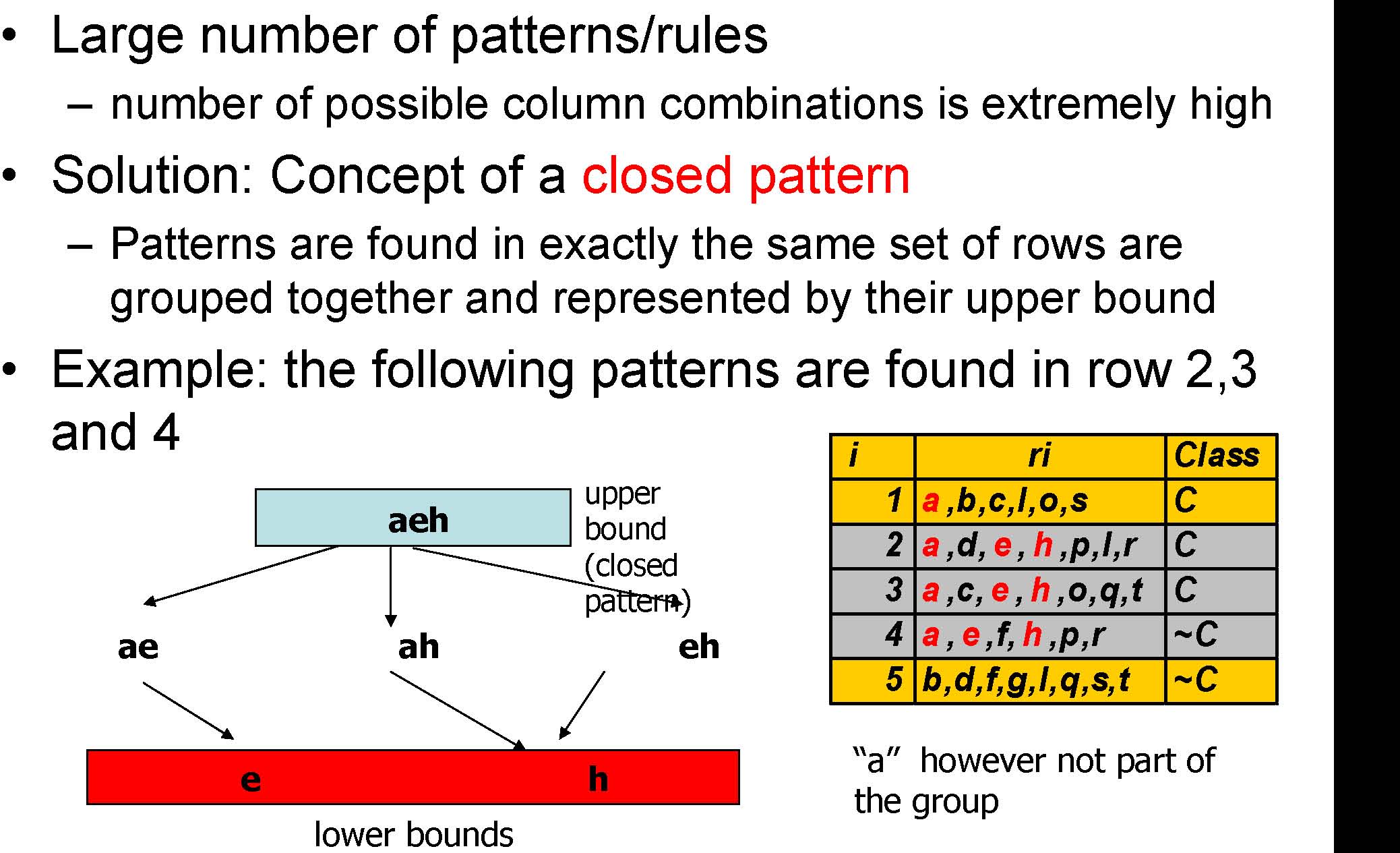
Challenge II
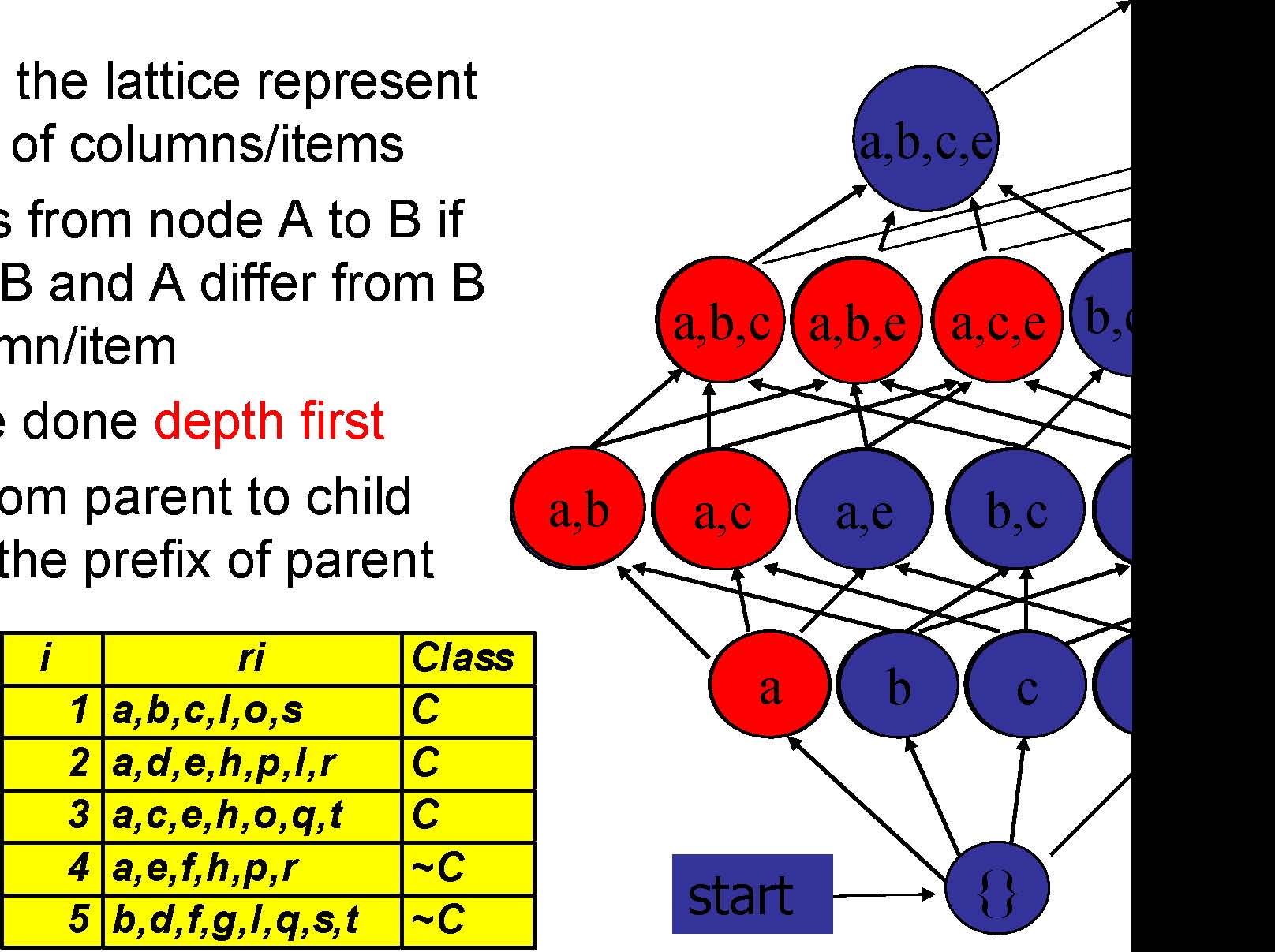
Column/Item Enumeration Lattice
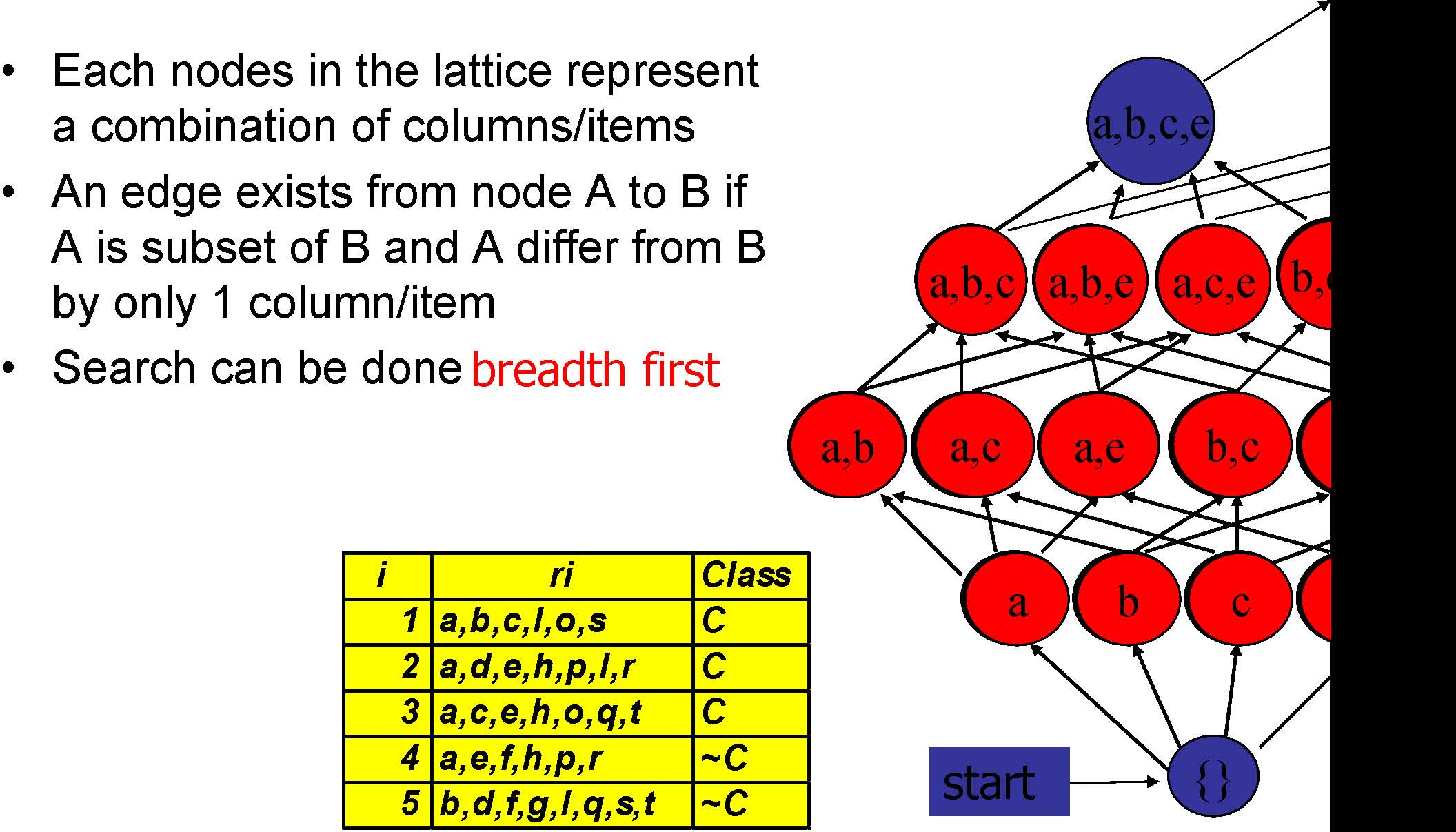
Column/Item Enumeration Lattice
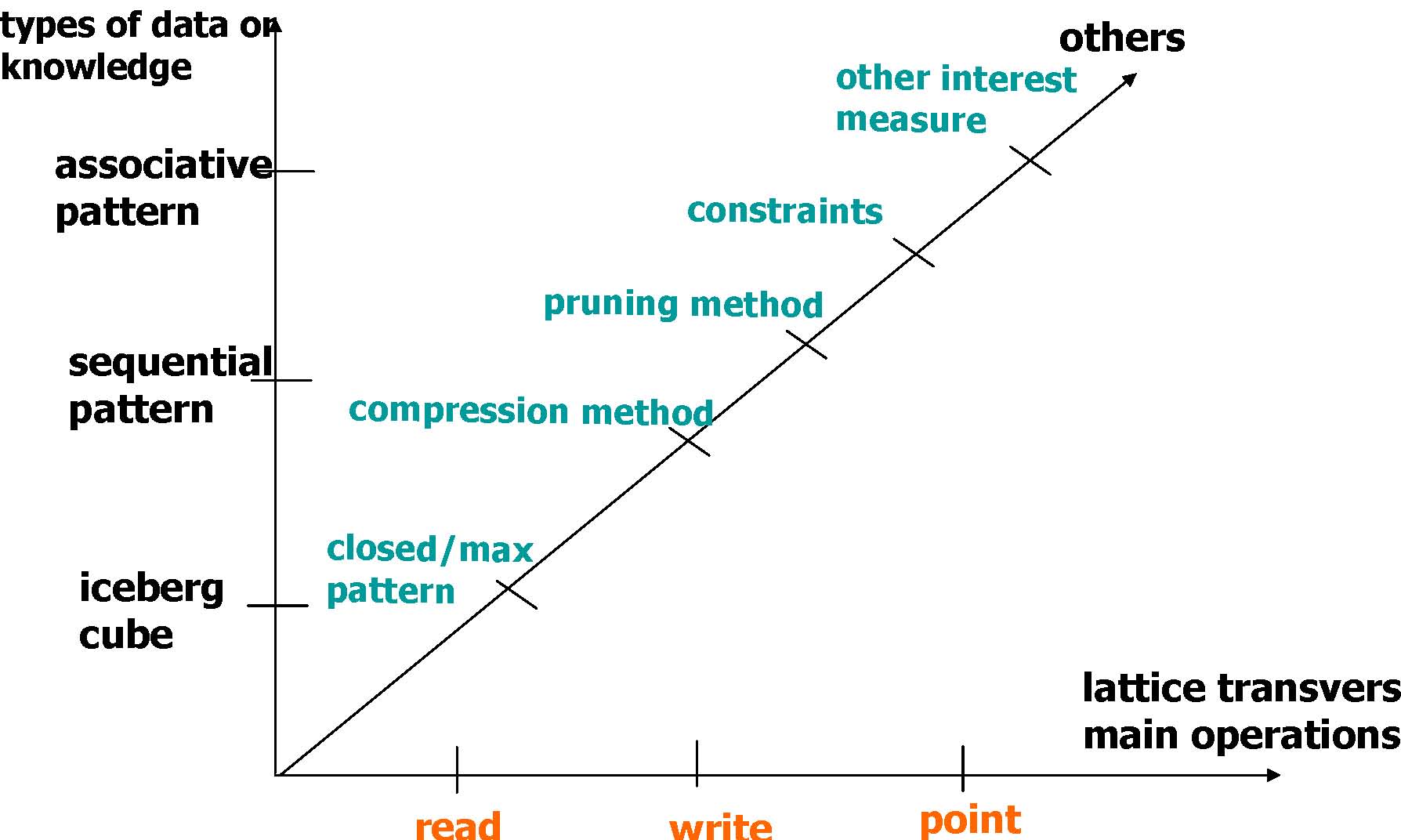
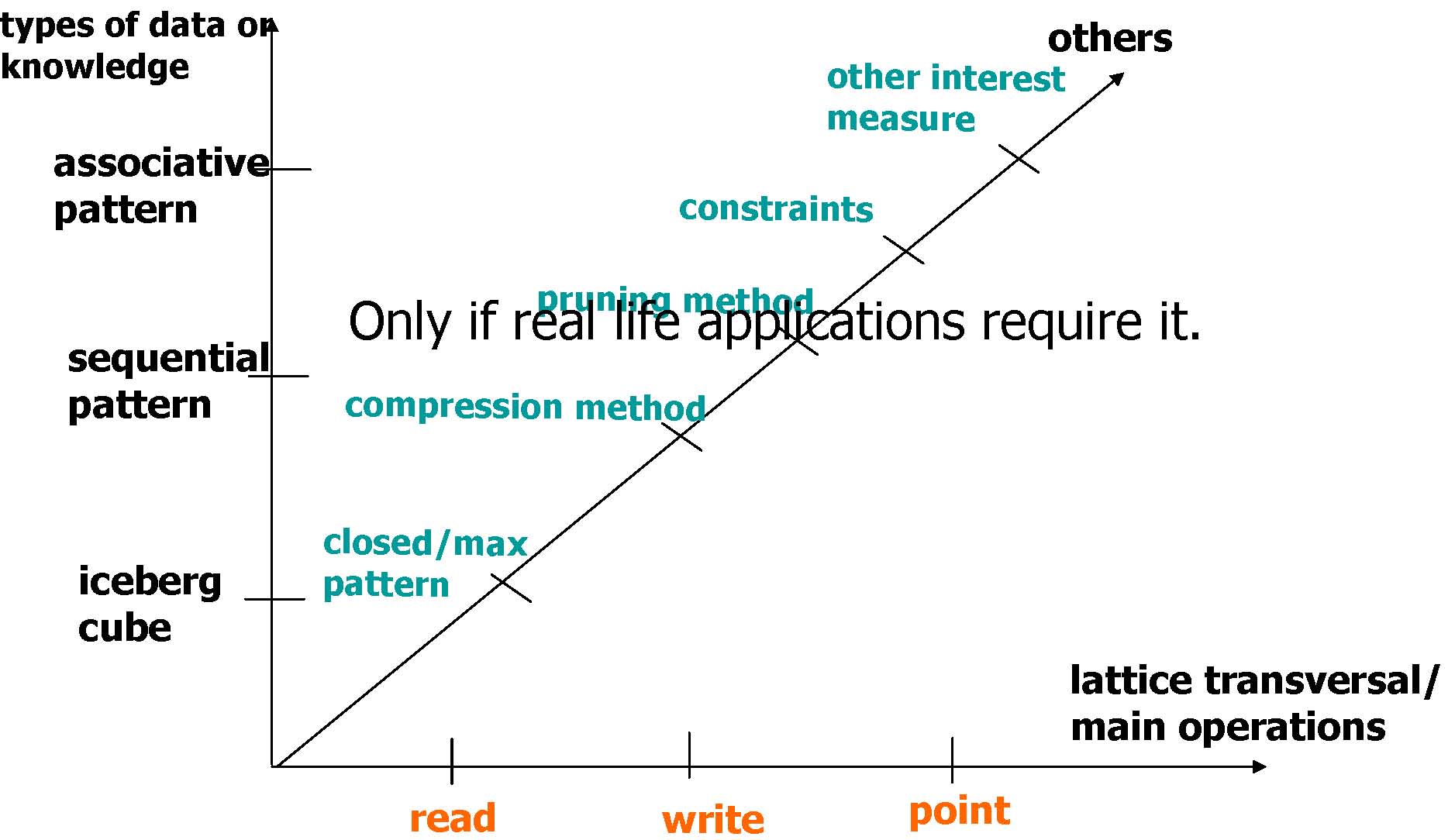
General Framework for Column/Item Enumeration
|
Read-based |
Write-based |
Point-based |
|
|
Association Mining |
Apriori[AgSr94], DIC |
Eclat, MaxClique[Zaki01], FPGrowth[HaPe00] |
Hmine |
|
Sequential Pattern Discovery |
GSP[AgSr96] |
SPADE [Zaki98,Zaki01], PrefixSpan[PHPC01] |
|
|
Iceberg Cube |
Apriori[AgSr94] |
BUC[BeRa99], H-Cubing [HPDW01] |
A Multidimensional View
read
write
Sample/Row Enumeration Algorihtms
Existing Sample Enumeration Algorithms
Concepts of CARPENTER
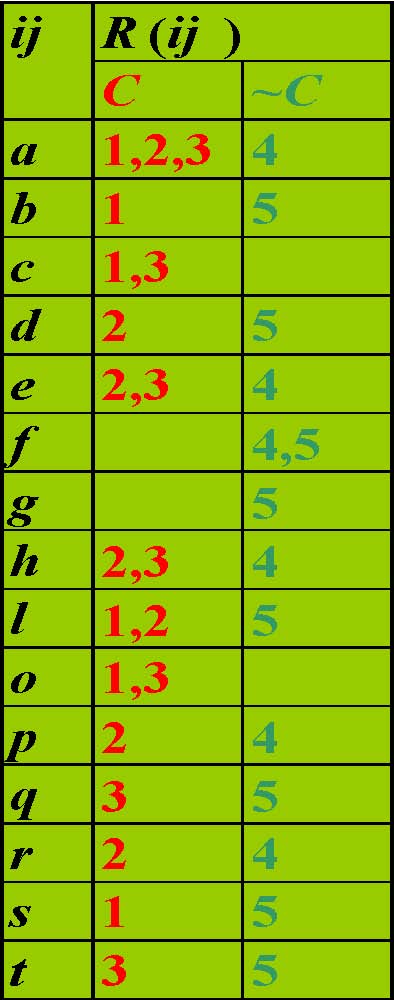
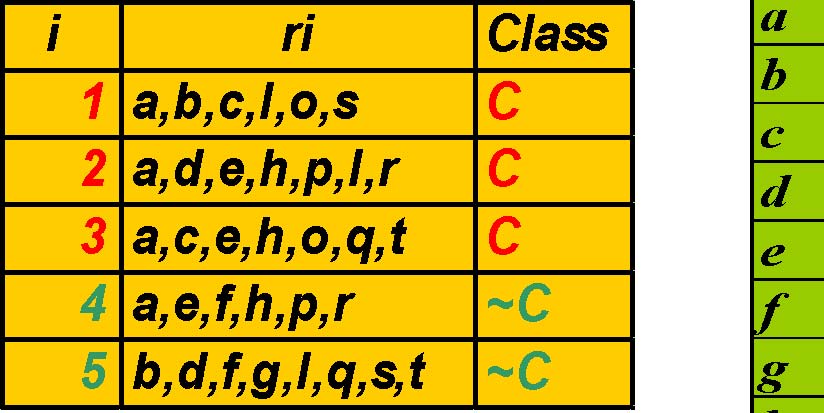
Example Table
Transposed Table,TT
TT|{2,3}

Row Enumeration
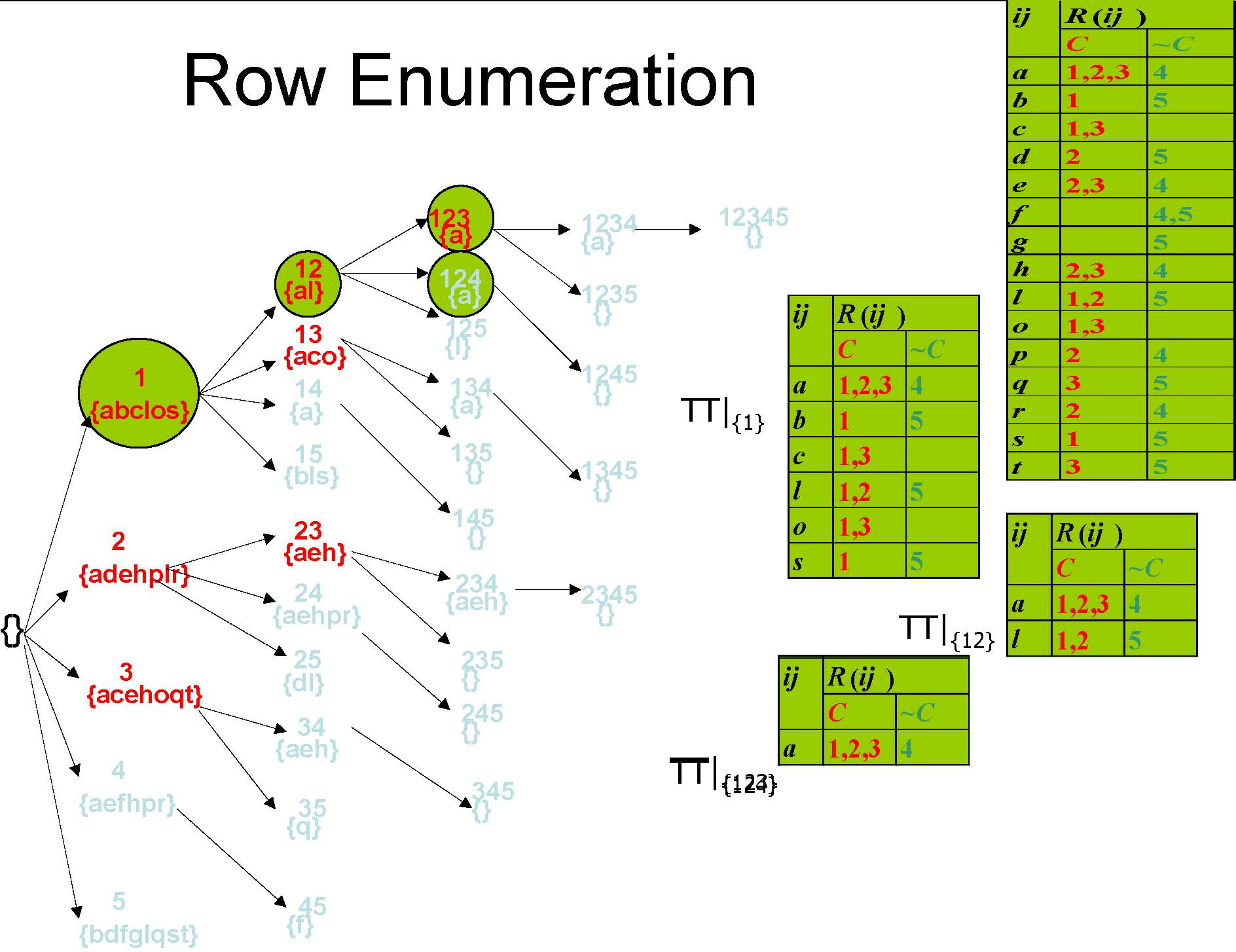
Pruning Method 1
r2 r3{aeh}
r4 has 100% support in the conditional table of “r2r3”, therefore branch “r2 r3r4”will be pruned.
r2 r3 r4 {aeh}
TT|{2,3}

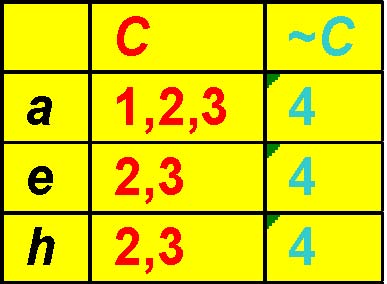
Pruning method 2
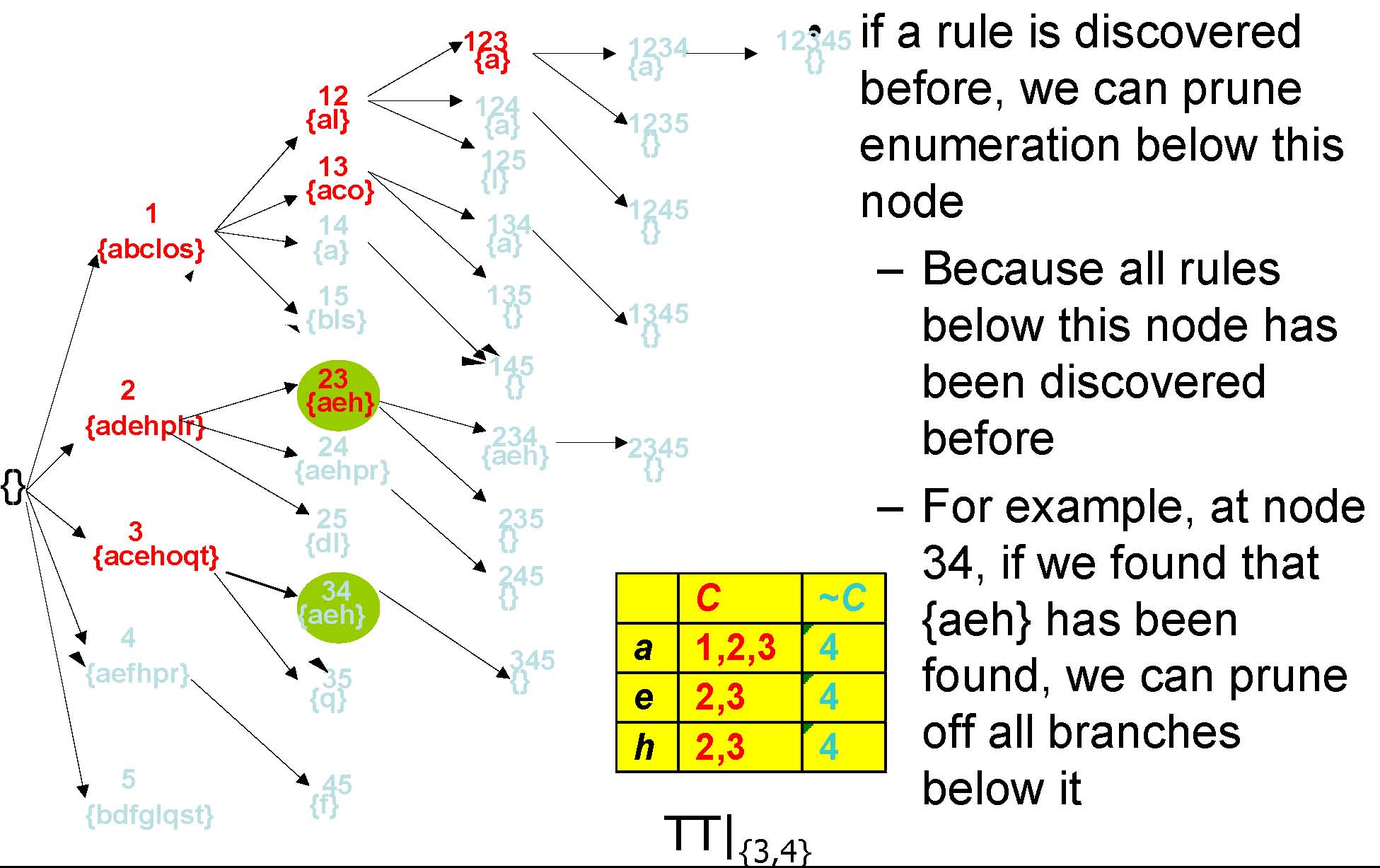
Pruning Method 3: Minimum Support

From CARPENTER to FARMER
Interesting Rule Groups
ae-->C (66%)
a-->C however is not in the group
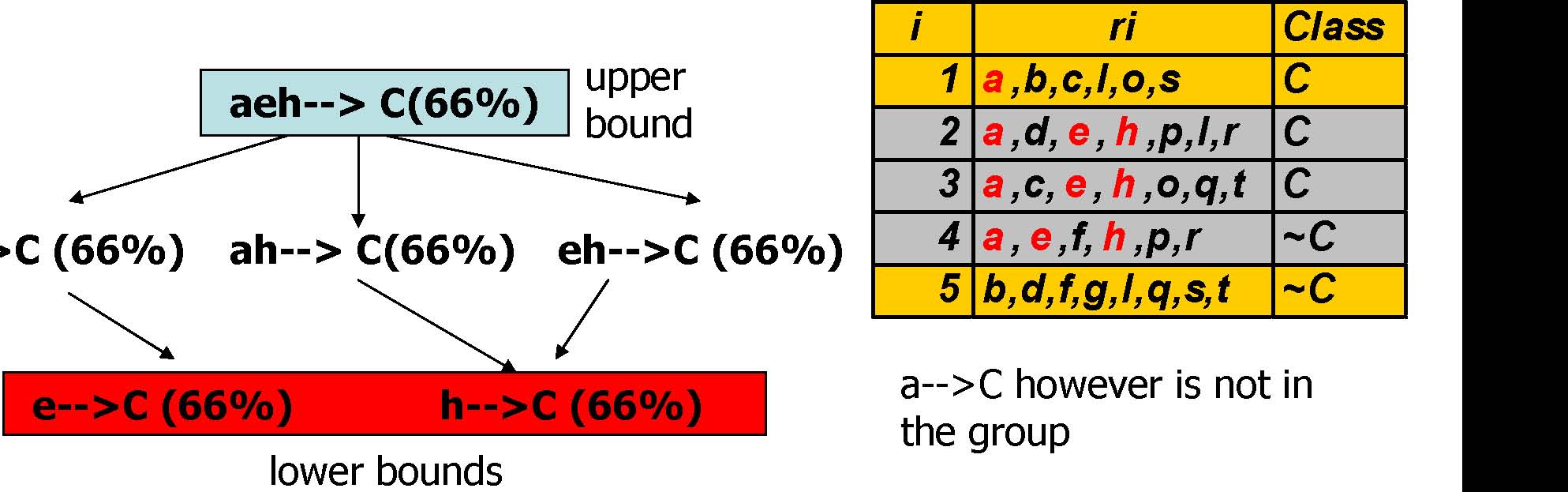
Pruning by Interestingness Measure
Ordering of Rows: All Class C before ~C
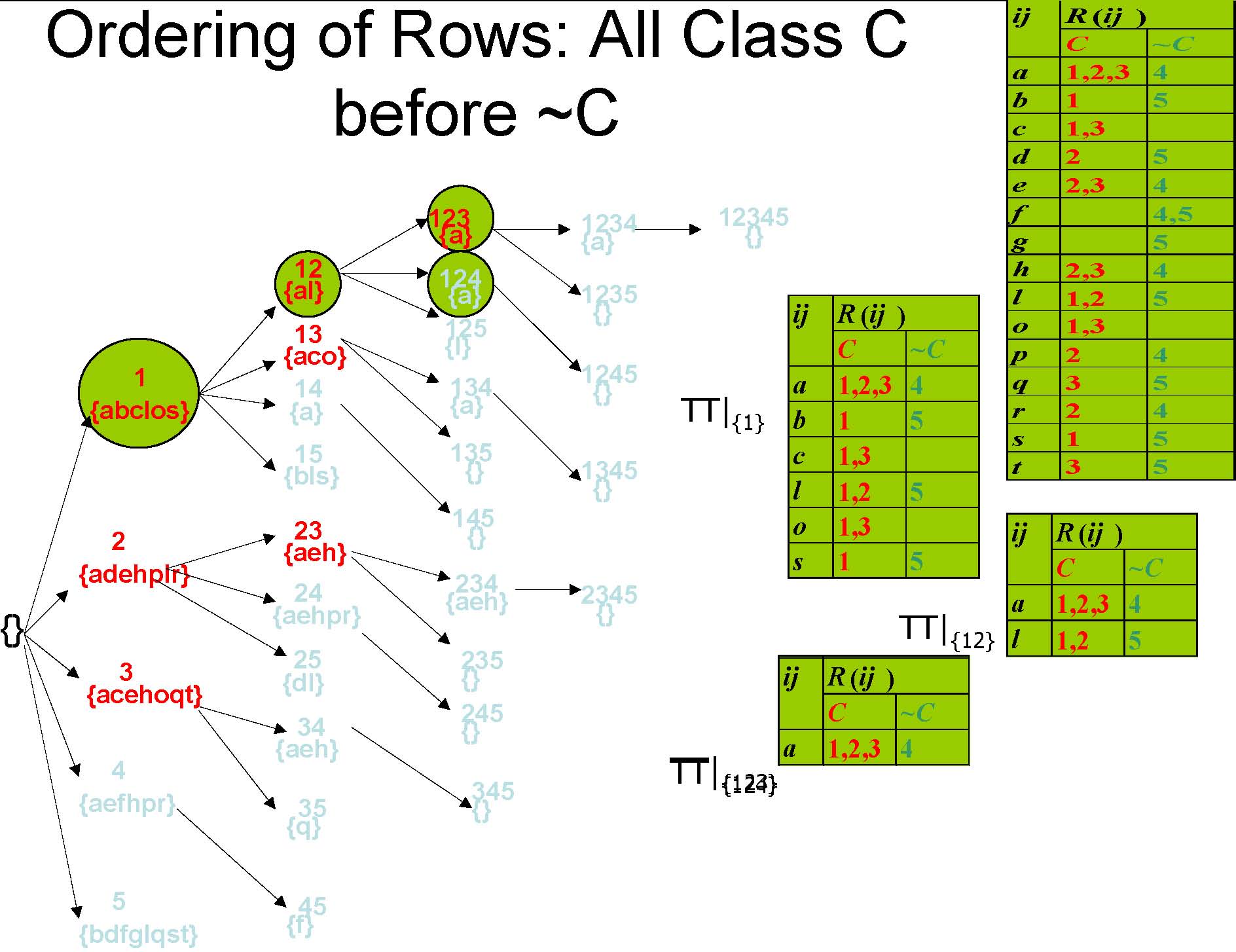
Pruning Method: Minimum Confidence
TT|{2,3}
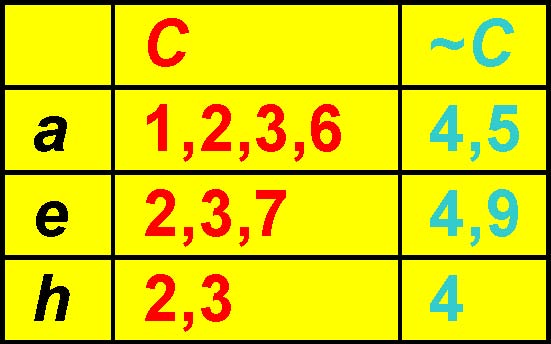
Pruning method: Minimum chi-square
TT|{2,3}
|
C |
~C |
Total |
|
|
A |
max=5 |
min=1 |
Computed |
|
~A |
Computed |
Computed |
Computed |
|
Constant |
Constant |
Constant |
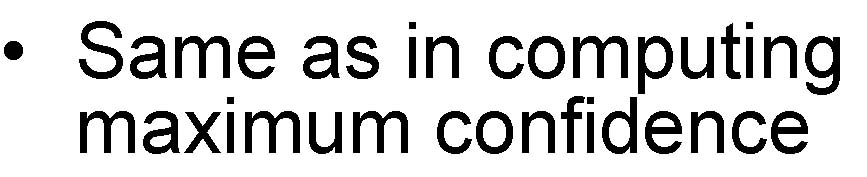
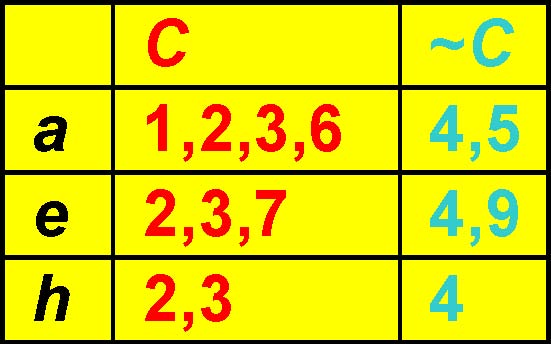
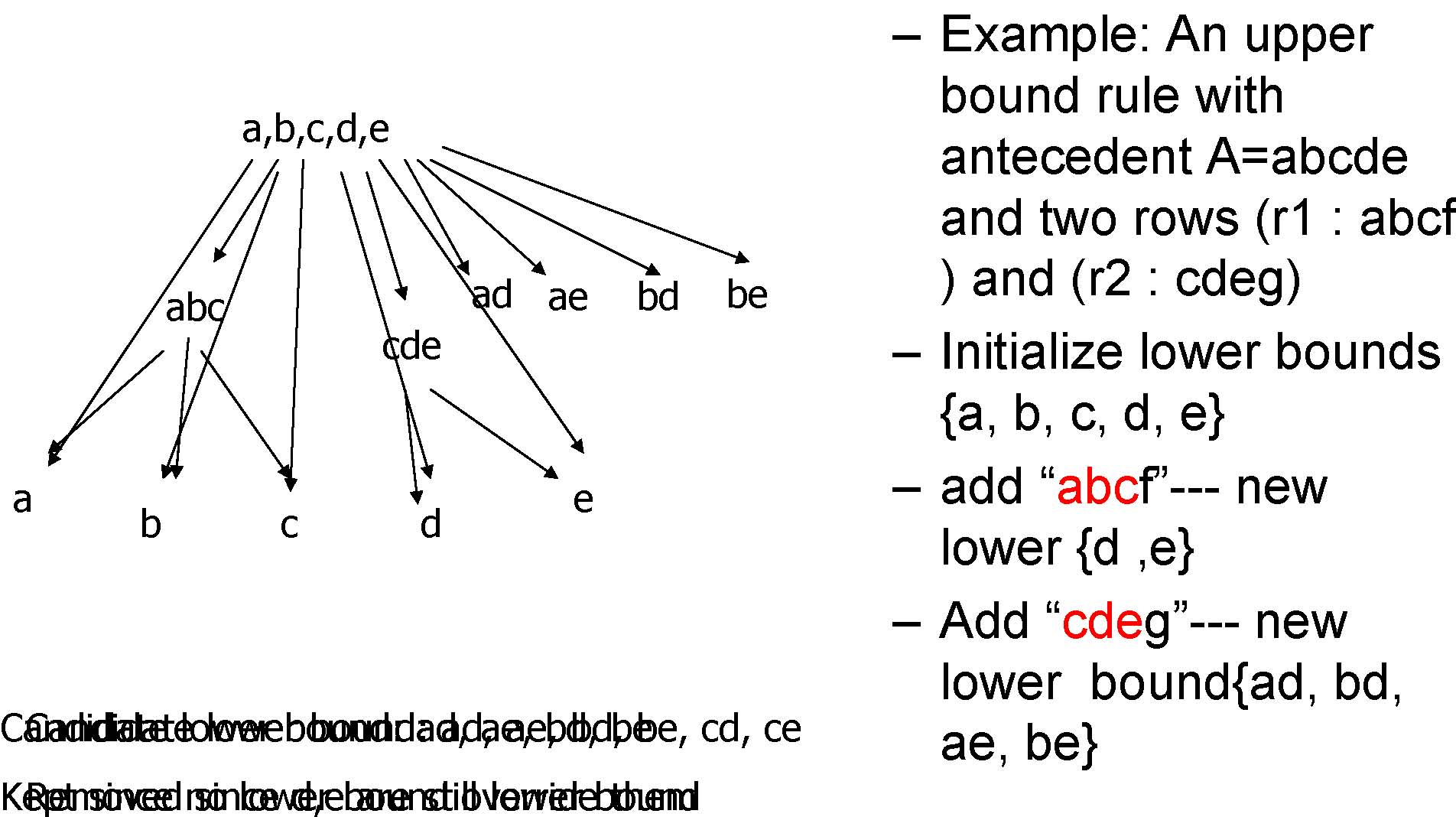
Finding Lower Bound, MineLB
Implementation

Experimental studies
Example results--Prostate
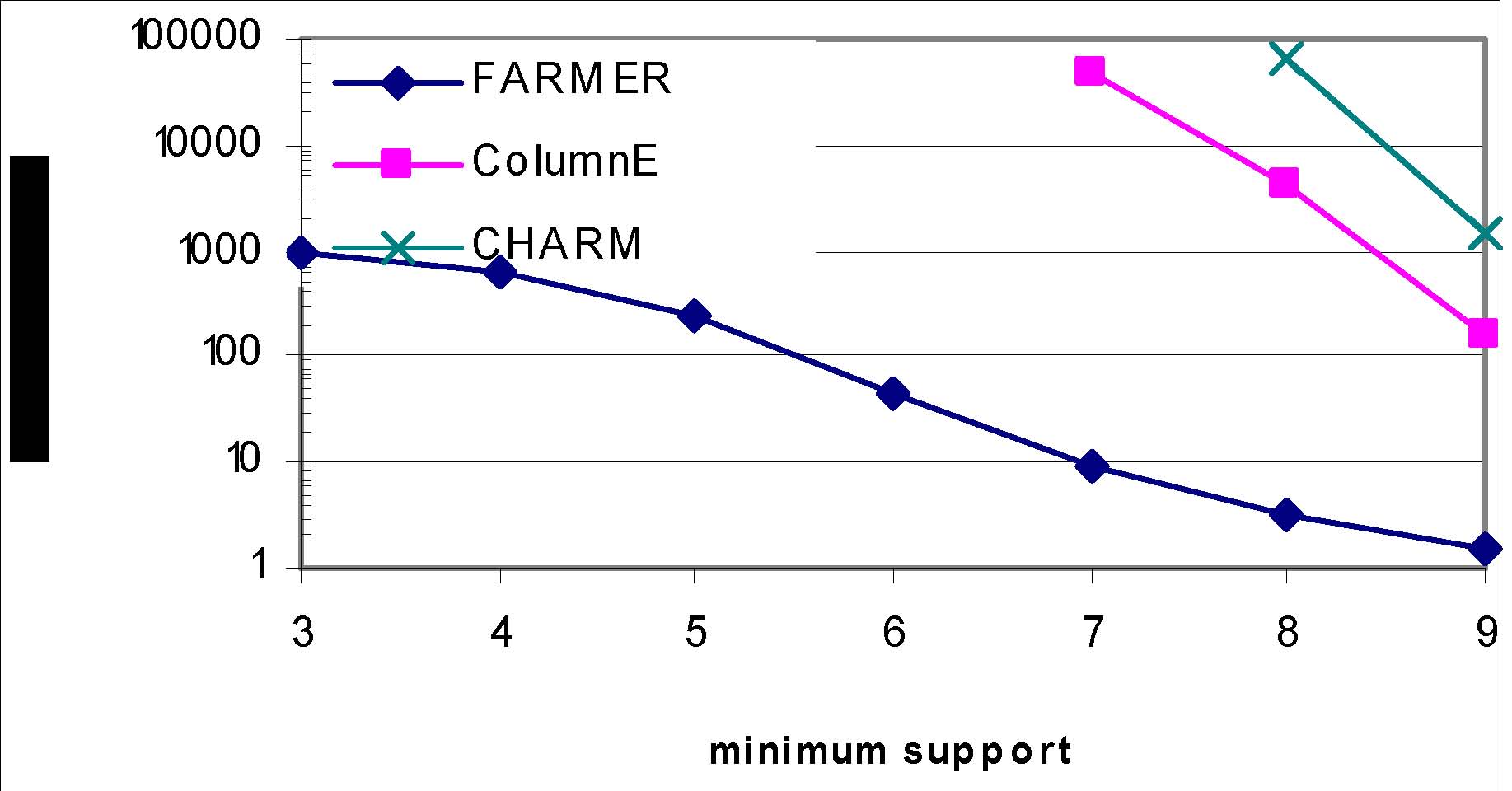
Example results--Prostate
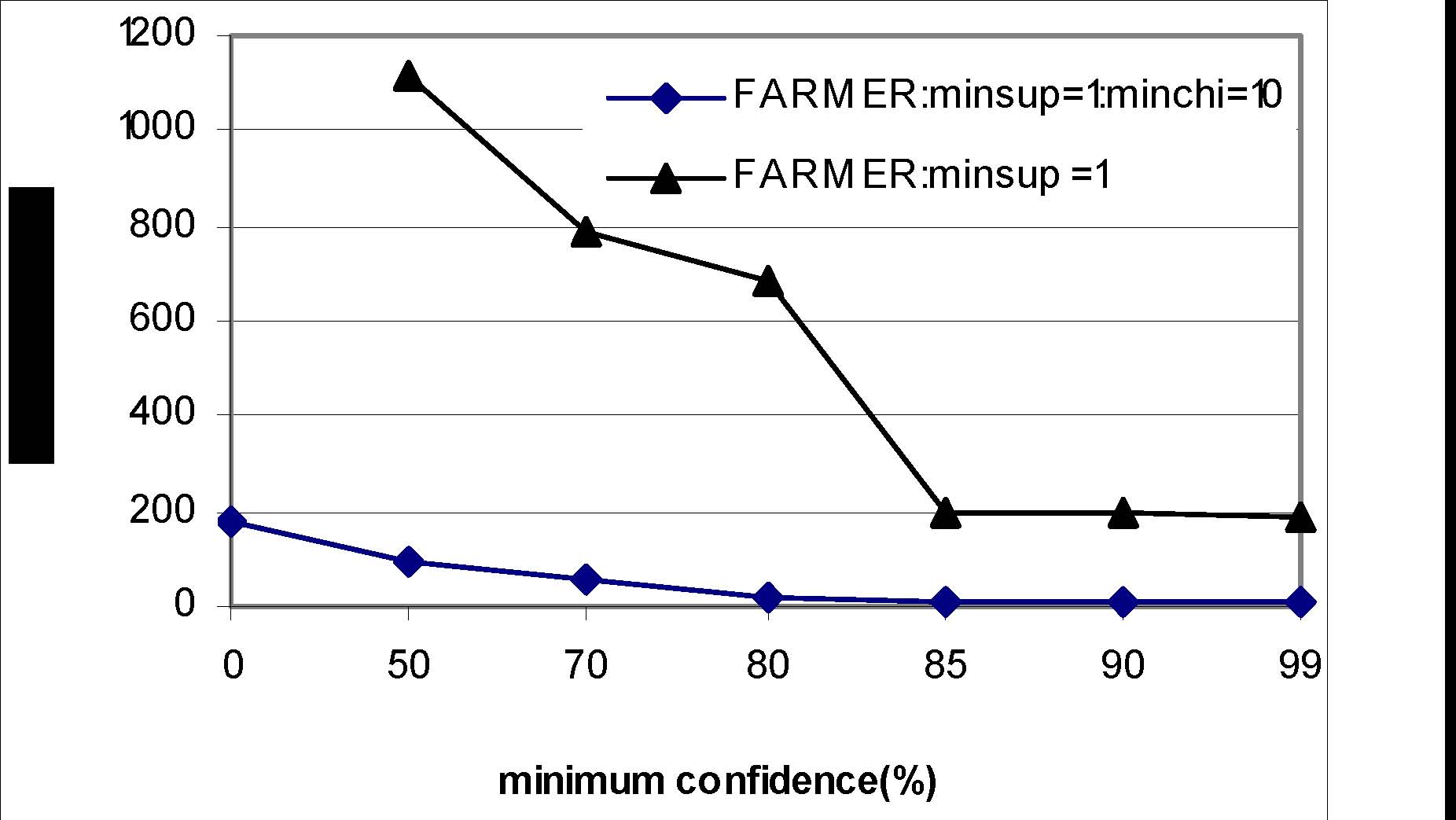
Naive Classification Approach
Classification results

Summary of Experiments
COBBLER: Combining Column and Row Enumeration
{ }
ab
{r1}
ad {r2}
acd{ r2}
bcd
{ }
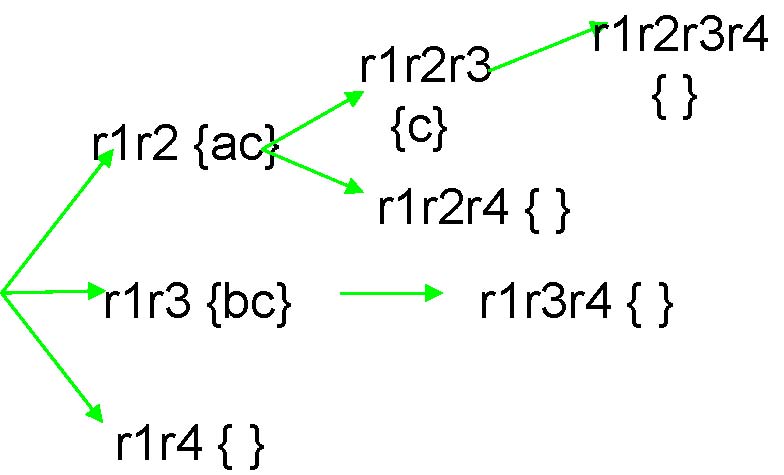
{ }
r1r3r4 { }
r2r4{d }
Single Enumeration Tree
Feature enumeration
Row enumeration
|
r1 |
a b c |
|
r2 |
a c d |
|
r3 |
b c |
|
r4 |
d |
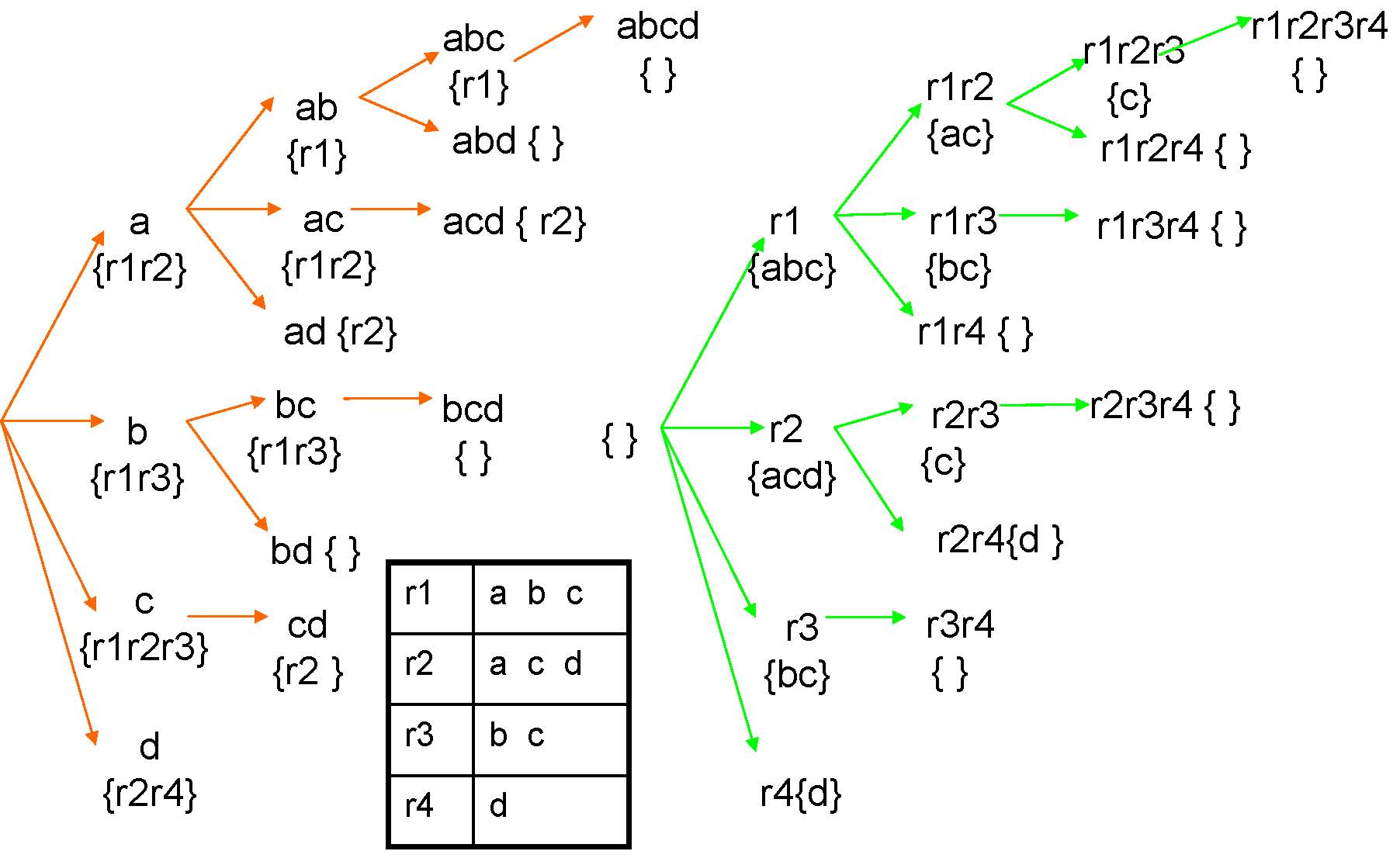
Dynamic Enumeration Tree
{ }
r1
{bc}
r1
{c}
r2
{d }
Feature enumerationto Row enumeration
ab
{r1}
abcd
{ }
a {r1r2}
abc: {r1}
ac: {r1r2}
acd: {r2}
|
r1 |
bc |
|
r2 |
cd |
|
br |
1 |
|
cr |
1 r2 |
|
dr |
2 |

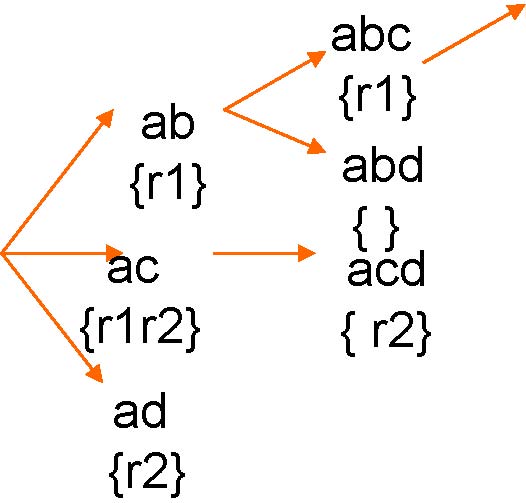
Dynamic Enumeration Tree
r4
{d}
c{r2r3 }
d {r4 }
b{r1 }
Row enumerationto Feature Enumeration
c{r1r2 }
bc{r1 }
r1 {abc}
r1r3r4 { }
ac: {r1r2}
bc: {r1r3}
c: {r1r2r3}

Switching Condition
Switching Condition


Switching Condition

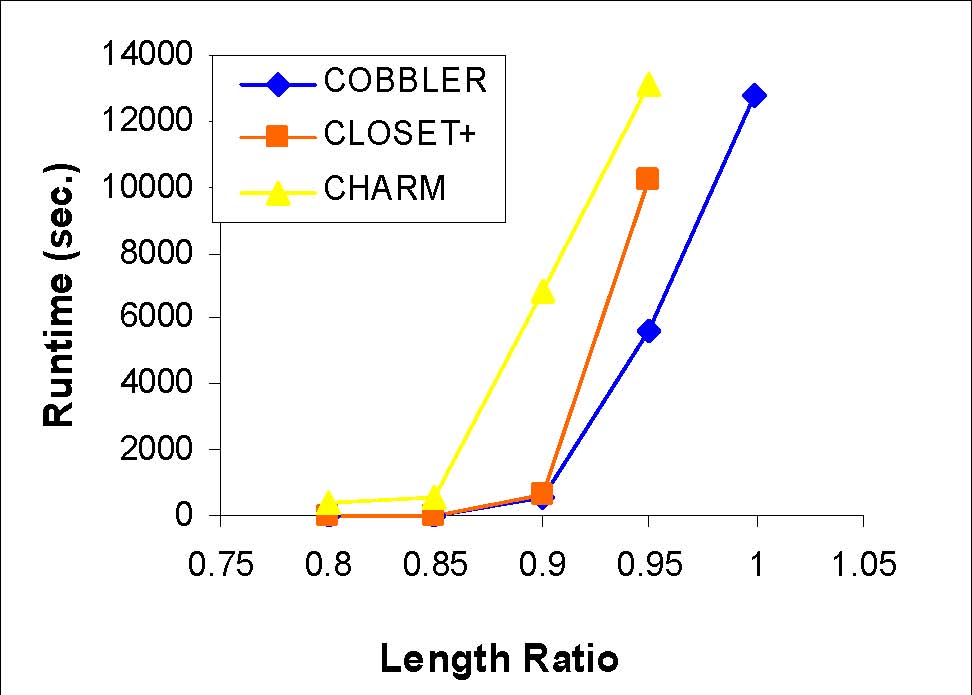
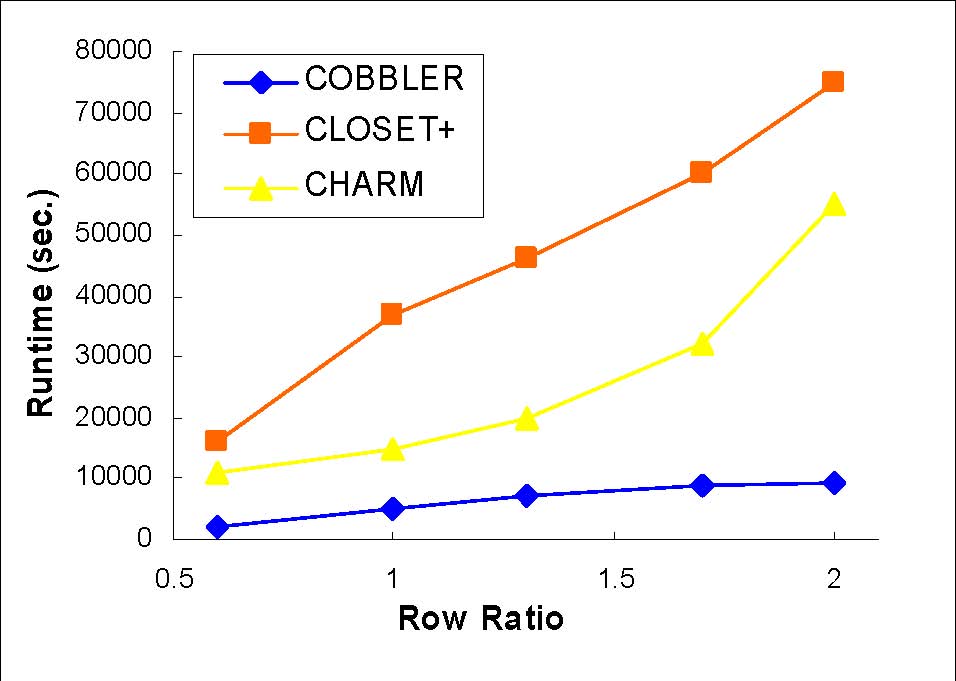
Length and Row ratio
Synthetic data
Extension of our work by other groups (with or without citation)
Extension of our work by other groups (with or without citation) II
|
Gene1 |
Gene2 |
Gene3 |
Gene4 |
|||||||
|
Sample1 |
||||||||||
|
Sample2 |
||||||||||
|
. . . |
||||||||||
|
SampleN-1 |
||||||||||
|
SampleN |
A gene in two samples are say to be coherent if their time series satisfied a certain matching condition
In CARPENTER, a gene in two samples are say to be matching if their expression in the two samples are almost the same
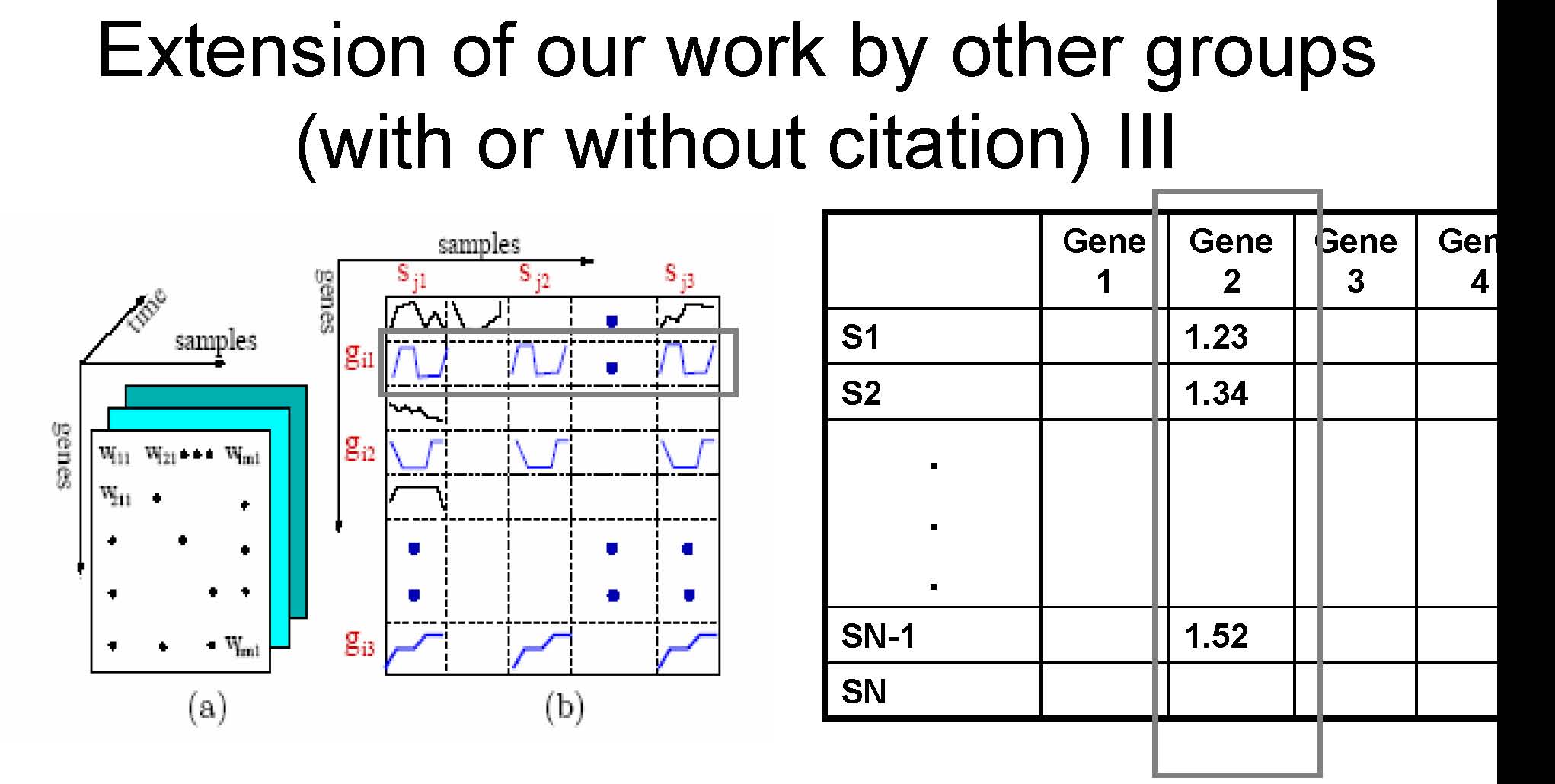
Extension of our work by other groups (with or without citation) IV
[2] Try to find a subset of samples S such that a subset of genes G is coherent for each pair of samples in S. |S|>mins, |G|>ming
In CARPENTER, we try to find a subset of samples S in which a subset of genes G is similar in expression level for each pair of samples in S. |S|>mins, |G|>0
|
Gene1 |
Gene2 |
Gene3 |
Gene4 |
|||||||
|
S1 |
1.23 |
|||||||||
|
S2 |
1.34 |
|||||||||
|
. . . |
||||||||||
|
SN-1 |
1.52 |
|||||||||
|
SN |
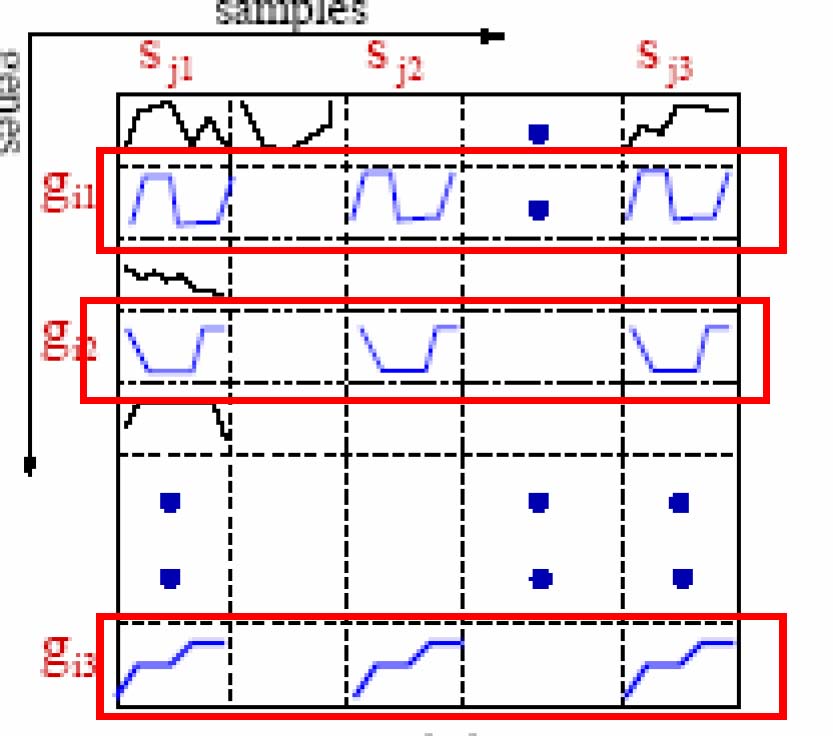
[2] Perform sample-wise enumeration and remove genes that are not pairwisecoherent across the samples enumerated
CARPENTER: Perform sample-wise enumeration and remove genes that does not have the same expression level across the samples enumerated
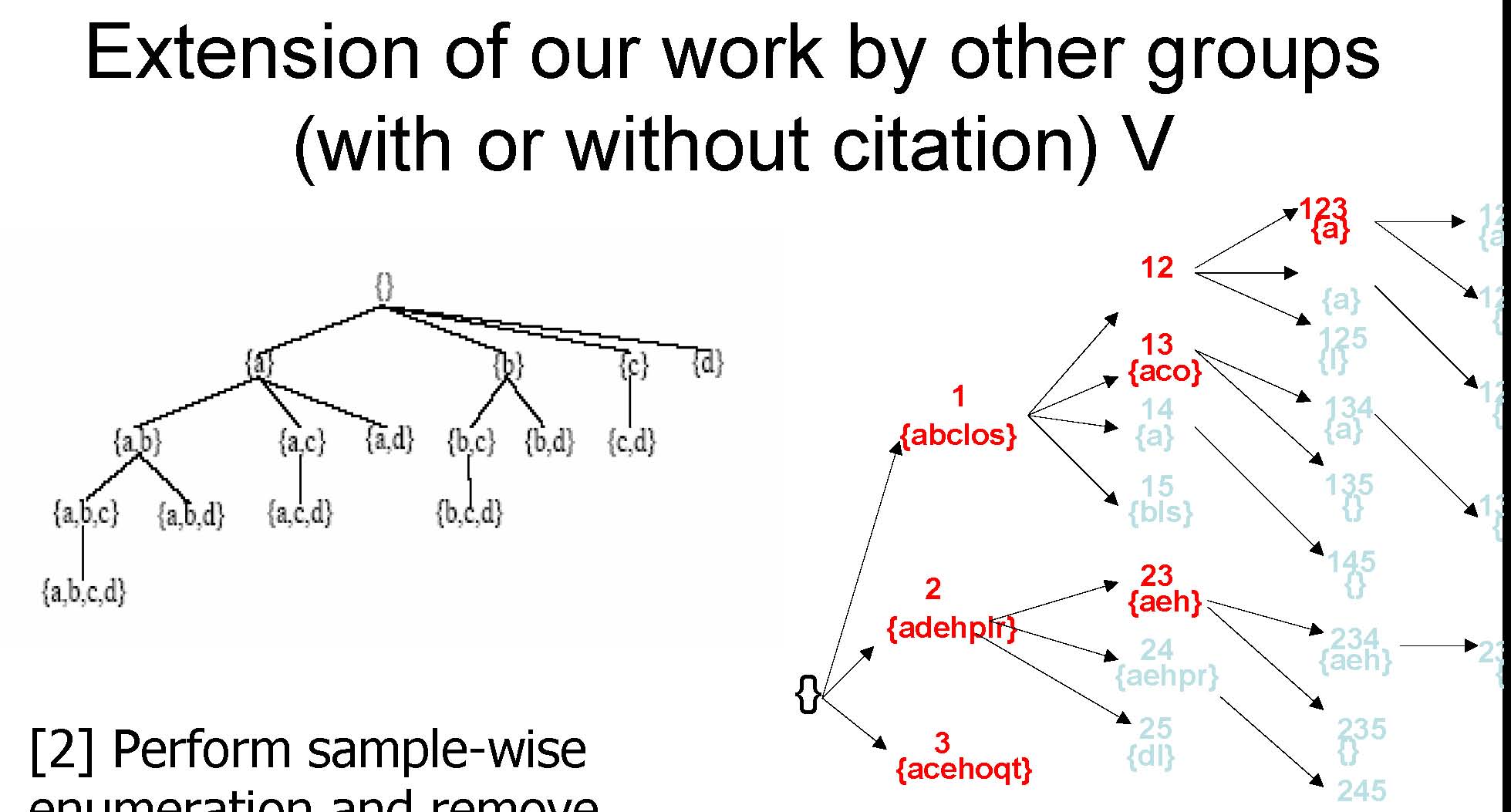
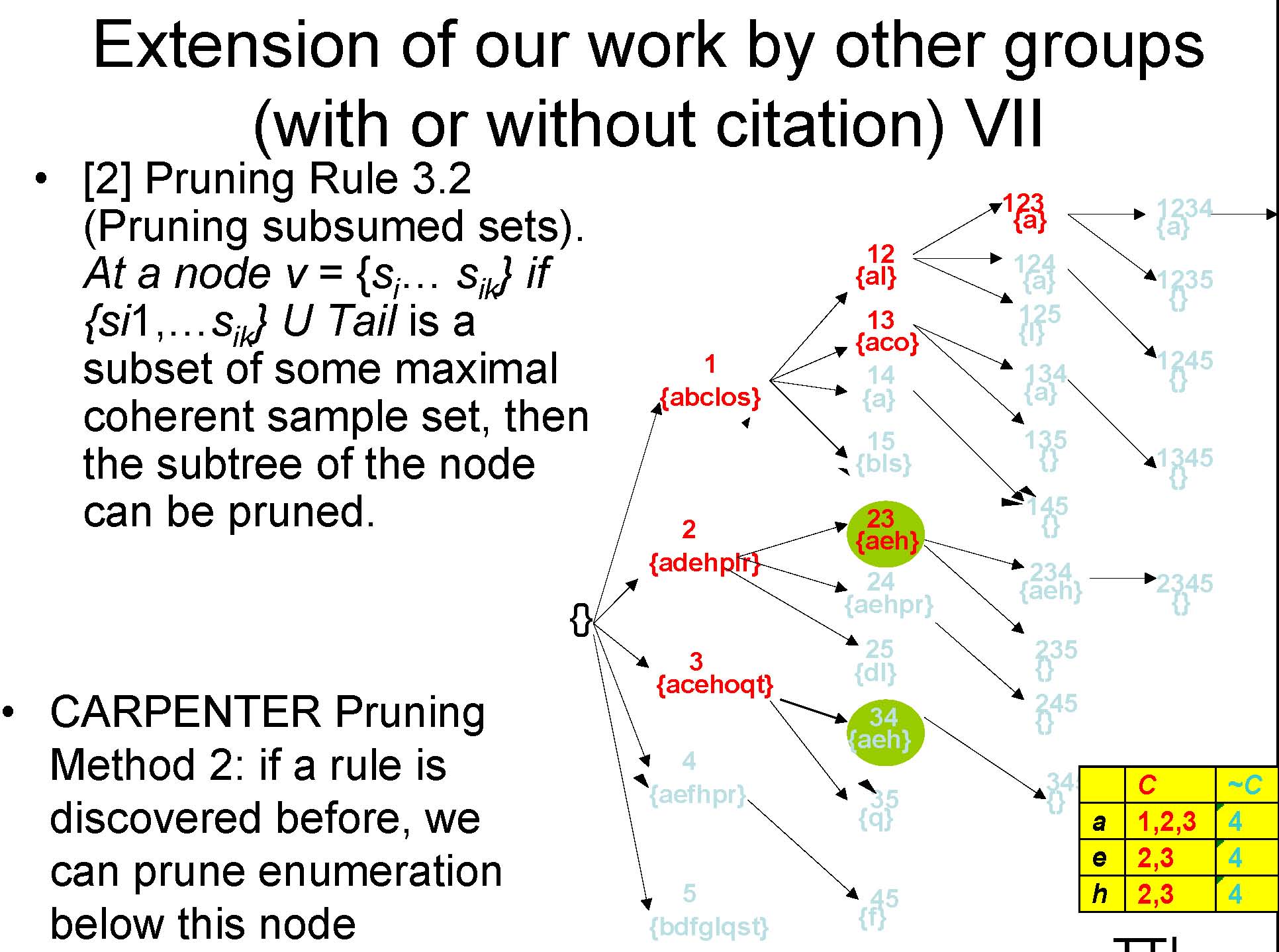
Extension of our work by other groups (with or without citation) VI
![Text Box: From [2]: Pruning Rule 3.1 (Pruning small sample sets). At a node v = fsi1 ; : : : ; sikg, the subtree of v can be pr](images/sample_enum2_img_41.jpg)
TT|{3,4}
Extension of our work (Conclusion)
Future Work: Generalize Framework for Row Enumeration Algorithms?
Conclusions