|
 Example alternative paths
Example alternative paths
|
|
In our previous works, we noted biological studies have showed that the interaction clusters obtained from contiguous connections forming closed loops in PPI networks have indicated an increased likelihood of biological relevance for the corresponding potential interactions.
Proteins that are found together within a circular contig in yeast-two-hybrid screens have been detected for known proteins in macromolecular complexes as well as signal transduction pathways. These observations have led to the use of alternative interaction paths in protein interaction networks as a measure to indicate the functional linkage between two proteins.
We illustrate here several actual examples from our yeast PPI dataset in which the presence or absence of alternative paths can be used to increase or decrease the confidence of protein-protein interactions.
|
|
 Example 1: Absence of or weak alternative path indicating a false positive PPI Example 1: Absence of or weak alternative path indicating a false positive PPI
|
|
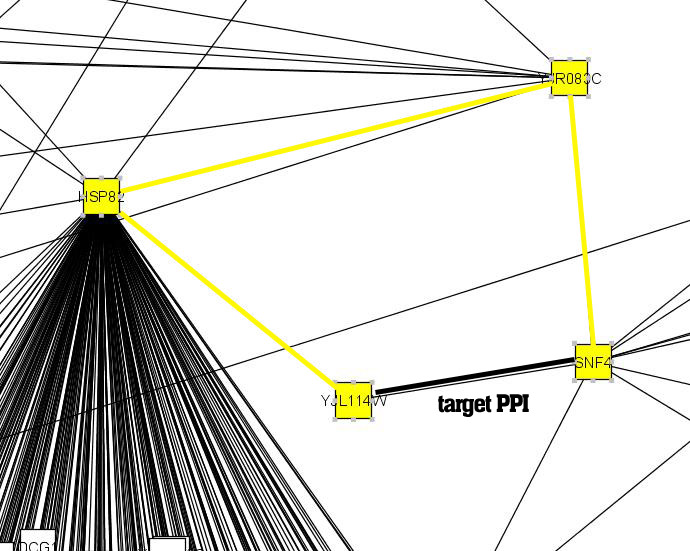
An interaction has been detected between the protein pair <Snf4, Yjl114w> (BIND ID 6321323 and 6322348) with Y2H assays. However, the degree of functional homogeneity between the pair of proteins, as measure by enriched GO term similarity, is as low as 0.062224. This biological observation indicates that the detected interaction between <Snf4, Yjl114w> is highly likely to be a false positive.
We verify whether we can come to the same conclusion using only network topological measures. The reversed IG1 value for <Snf4, Yjl114w> is a high 0.977012, which supports the possibly wrong suggestion that this is a true interaction. In contrast, the IRAP value is a low 0.02108, which means that even the strongest alternative path between the two proteins <Snf4, Yjl114w> (in this case, "Snf4-Yjr083c-Hsp82-Yjl114w") has
been deemed unreliable in our IRAP model. This concurs well with the biological observation of a low degree of functional homogeneity between the proteins.
Biological evidence: the related GO terms for the target protein pair are:
SGD S000003083 SNF4 GO:0005634 SGD_REF:S000046049|PMID:2481228 IDA C associates with Snf1p YGL115W|CAT3|SCI1 gene taxon:4932 20020806 SGD
SGD S000003083 SNF4 GO:0005737 SGD_REF:S000046049|PMID:2481228 IDA C associates with Snf1p YGL115W|CAT3|SCI1 gene taxon:4932 20020806 SGD
SGD S000003083 SNF4 GO:0005886 SGD_REF:S000072977|PMID:12562756 IDA C associates with Snf1p YGL115W|CAT3|SCI1 gene taxon:4932 20030425 SGD
SGD S000003083 SNF4 GO:0030295 SGD_REF:S000053604|PMID:2557546 IGI F associates with Snf1p YGL115W|CAT3|SCI1 gene taxon:4932 20020806 SGD
SGD S000003083 SNF4 GO:0006357 SGD_REF:S000053604|PMID:2557546 IGI P associates with Snf1p YGL115W|CAT3|SCI1 gene taxon:4932 20020806 SGD
SGD S000003083 SNF4 GO:0007031 SGD_REF:S000047076|PMID:1355328 IMP P associates with Snf1p YGL115W|CAT3|SCI1 gene taxon:4932 20020806 SGD
SGD S000003650 YJL114W GO:0000943 SGD_REF:S000072465|PMID:9582191 ISS C YJL114W gene taxon:4932 20030811 SGD
SGD S000003650 YJL114W GO:0003723 SGD_REF:S000072465|PMID:9582191 ISS F YJL114W gene taxon:4932 20030811 SGD
SGD S000003650 YJL114W GO:0005515 SGD_REF:S000072465|PMID:9582191 ISS F YJL114W gene taxon:4932 20030811 SGD
SGD S000003650 YJL114W GO:0006319 SGD_REF:S000072465|PMID:9582191 ISS P YJL114W gene taxon:4932 20030811 SGD
|
|
 Example 2: Strong alternative path indicating a true positive PPI Example 2: Strong alternative path indicating a true positive PPI
|
|
In the previous example, a low IRAP value indicates a false positive PPI. IRAP can thus be used to detect and eliminate possible false positives in an interactome. In IRAP*, we need to detect true positives in addition of identifying the possible false positives for removal. To detect the true positives, the presence of a strong alternative path should indicate a true positive PPI. We give two examples below: the first example illustrates that IRAP can detect
the same true positive as IG1, while the second example shows a PPI that was missed by IG1 but was detected with our IRAP model.
|
|
 Example 2.1: IRAP is high, IG1 is high Example 2.1: IRAP is high, IG1 is high
|
|
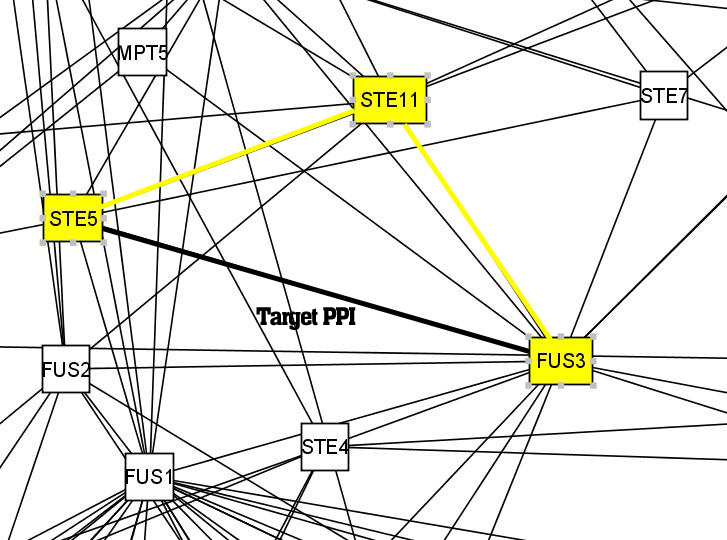
The protein pair <Ste5, Fus3> (BIND ID 6320308 and 6319455, labeled as 5 and 32 in the following figure) has a high degree of functional homogeneity - in fact, its enriched GO term similarity is 1.000000. This interaction has a high probability to be true, because the two proteins have the same functions (MAP-kinase scaffold activity, signal transduction during conjugation with cellular fusion, etc.,) and are located at the same place (e.g. cytoplasm) in the cell.
We verify whether using only network topological information can also help us identify this interaction. Indeed, both its IRAP and reversed IG1 values are 1.0000. The alternative path selected by IRAP was "Ste5-Ste11-Fus3". In this case, both IRAP and IG1 correctly identified the true positive PPI.
Biological evidence: the related GO terms for the target protein pair are
SGD S000002510 STE5 GO:0005634 SGD_REF:S000053527|PMID:9732267 IDA C YDR103W|HMD3|NUL3 gene taxon:4932 20010118 SGD
SGD S000002510 STE5 GO:0005737 SGD_REF:S000069153|PMID:11781566 IDA C YDR103W|HMD3|NUL3 gene taxon:4932 20020829 SGD
SGD S000002510 STE5 GO:0005886 SGD_REF:S000053527|PMID:9732267 IDA C YDR103W|HMD3|NUL3 gene taxon:4932 20010118 SGD
SGD S000002510 STE5 GO:0043332 SGD_REF:S000069153|PMID:11781566 IDA C YDR103W|HMD3|NUL3 gene taxon:4932 20020829 SGD
SGD S000002510 STE5 GO:0005078 SGD_REF:S000050302|PMID:7851759 IPI F YDR103W|HMD3|NUL3 gene taxon:4932 20010118 SGD
SGD S000002510 STE5 GO:0005078 SGD_REF:S000051962|PMID:8062390 IPI F YDR103W|HMD3|NUL3 gene taxon:4932 20020403 SGD
SGD S000002510 STE5 GO:0000750 SGD_REF:S000051962|PMID:8062390 IDA P YDR103W|HMD3|NUL3 gene taxon:4932 20021206 SGD
SGD S000002510 STE5 GO:0000750 SGD_REF:S000051962|PMID:8062390 IPI P YDR103W|HMD3|NUL3 gene taxon:4932 20021206 SGD
SGD S000002510 STE5 GO:0001403 SGD_REF:S000082049|PMID:15192700 IMP P YDR103W|HMD3|NUL3 gene taxon:4932 20050930 SGD
SGD S000000112 FUS3 GO:0005634 SGD_REF:S000048143|PMID:10233162 IDA C CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20010118 SGD
SGD S000000112 FUS3 GO:0005737 SGD_REF:S000048143|PMID:10233162 IDA C CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20010118 SGD
SGD S000000112 FUS3 GO:0005739 SGD_REF:S000075100|PMID:14576278 IDA C CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20041203 SGD
SGD S000000112 FUS3 GO:0043332 SGD_REF:S000069153|PMID:11781566 IDA C CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20020829 SGD
SGD S000000112 FUS3 GO:0004707 SGD_REF:S000045748|PMID:8384702 TAS F CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20010118 SGD
SGD S000000112 FUS3 GO:0004707 SGD_REF:S000080086|PMID:15620357 IDA F CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20050218 SGD
SGD S000000112 FUS3 GO:0000750 SGD_REF:S000045748|PMID:8384702 IDA P CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20021206 SGD
SGD S000000112 FUS3 GO:0001402 SGD_REF:S000039888|PMID:9393860 IMP P CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20050218 SGD
SGD S000000112 FUS3 GO:0001403 SGD_REF:S000050425|PMID:8001818 IMP P CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20050218 SGD
SGD S000000112 FUS3 GO:0001403 SGD_REF:S000080086|PMID:15620357 IMP P CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20050218 SGD
SGD S000000112 FUS3 GO:0001403 SGD_REF:S000062980|PMID:10652102 IMP P CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20050218 SGD
SGD S000000112 FUS3 GO:0006468 SGD_REF:S000045748|PMID:8384702 TAS P CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20010118 SGD
SGD S000000112 FUS3 GO:0007050 SGD_REF:S000055079|PMID:10049917 TAS P CDC28/cdc2 related protein kinase YBL016W|DAC2 gene taxon:4932 20010118 SGD
|
|
 Example 2.2: IRAP is high, IG1 is low Example 2.2: IRAP is high, IG1 is low
|
|
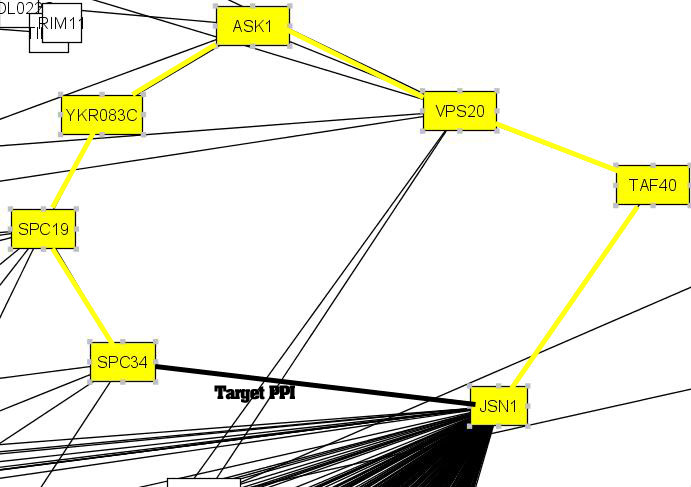
Another protein pair of interest is <Spc34, Jsn1> (BIND ID 6322890 and 6322550). This interaction was supported by the high degree of functional homogeneity between the two proteins, which have a relatively high enriched GO term similarity of 0.886994.
For this interaction, IG1 failed to detect it. The reversed IG1 value is a low 0.103448. On the other hand, it has a relatively high IRAP value of 0.504180, with an alternative path of "Spc34-Spc19-Ykr083c-Ask1-Vps20-Taf40-Jsn1". Note that in this case, the corresponding alternative path detected by IRAP is fairly long, illustrating that the shortest path need not be the strongest one.
Biological evidence: the related GO terms for the target protein pair are
SGD S000001745 SPC34 GO:0000778 SGD_REF:S000069091|PMID:11756468 IDA C spindle pole component YKR037C gene taxon:4932 20021008 SGD
SGD S000001745 SPC34 GO:0000778 SGD_REF:S000080960|PMID:11511347 IDA C spindle pole component YKR037C gene taxon:4932 20050307 SGD
SGD S000001745 SPC34 GO:0005816 SGD_REF:S000047788|PMID:9585415 IDA C spindle pole component YKR037C gene taxon:4932 20010118 SGD
SGD S000001745 SPC34 GO:0005819 SGD_REF:S000069091|PMID:11756468 IDA C spindle pole component YKR037C gene taxon:4932 20021008 SGD
SGD S000001745 SPC34 GO:0005876 SGD_REF:S000080960|PMID:11511347 IDA C spindle pole component YKR037C gene taxon:4932 20050307 SGD
SGD S000001745 SPC34 GO:0042729 SGD_REF:S000069052|PMID:11799062 IDA C spindle pole component YKR037C gene taxon:4932 20040122 SGD
SGD S000001745 SPC34 GO:0042729 SGD_REF:S000080667|PMID:15664196 IDA C spindle pole component YKR037C gene taxon:4932 20050328 SGD
SGD S000001745 SPC34 GO:0042729 SGD_REF:S000080444|PMID:15640796 IPI C spindle pole component YKR037C gene taxon:4932 20050328 SGD
SGD S000001745 SPC34 GO:0042729 SGD_REF:S000080444|PMID:15640796 IDA C spindle pole component YKR037C gene taxon:4932 20050328 SGD
SGD S000001745 SPC34 GO:0042729 SGD_REF:S000069052|PMID:11799062 IPI C spindle pole component YKR037C gene taxon:4932 20040122 SGD
SGD S000001745 SPC34 GO:0042729 SGD_REF:S000080667|PMID:15664196 IPI C spindle pole component YKR037C gene taxon:4932 20050328 SGD
SGD S000001745 SPC34 GO:0005200 SGD_REF:S000047788|PMID:9585415 IPI F spindle pole component YKR037C gene taxon:4932 20010118 SGD
SGD S000001745 SPC34 GO:0007020 SGD_REF:S000058447|PMID:9153752 IPI P spindle pole component YKR037C gene taxon:4932 20010118 SGD
SGD S000001745 SPC34 GO:0030472 SGD_REF:S000069091|PMID:11756468 IPI P spindle pole component YKR037C gene taxon:4932 20021008 SGD
SGD S000001745 SPC34 GO:0031110 SGD_REF:S000080667|PMID:15664196 IPI P spindle pole component YKR037C gene taxon:4932 20050328 SGD
SGD S000001745 SPC34 GO:0031110 SGD_REF:S000080667|PMID:15664196 IDA P spindle pole component YKR037C gene taxon:4932 20050328 SGD
SGD S000003851 JSN1 GO:0008372 SGD_REF:S000069584 ND C YJR091C|PUF1 gene taxon:4932 20020924 SGD
SGD S000003851 JSN1 GO:0003729 SGD_REF:S000059700|PMID:11101532 ISS F YJR091C|PUF1 gene taxon:4932 20020924 SGD
SGD S000003851 JSN1 GO:0000288 SGD_REF:S000059700|PMID:11101532 IGI P YJR091C|PUF1 gene taxon:4932 20020924 SGD
|
|













