
Yang ZHANG
ProfessorDepartment of Computer Science, the School of Computing
Department of Biochemistry, Yong Loo Lin School of Medicine
Cancer Science Institute of Singapore
Dr Yang Zhang is a Professor in the Department of Computer Science at the School of Computing, National University of Singapore (NUS). He also holds joint appointments as a Professor in the Department of Biochemistry at the Yong Loo Lin School of Medicine, NUS, and at the Cancer Science Institute of Singapore. Prior to joining NUS, Dr Zhang was a Professor at the University of Michigan, with appointments in the Department of Computational Medicine and Bioinformatics, the Department of Biological Chemistry, and the Department of Macromolecular Science and Engineering. His research focuses on artificial intelligence (AI) and machine learning for biomolecular modeling, including protein and RNA folding and structure prediction, as well as protein, RNA and peptide sequence design. The I-TASSER algorithm (https://zhanggroup.org/I-TASSER/), developed in his laboratory, has been consistently ranked among the most accurate automated protein structure prediction methods in the community-wide CASP experiments over the past two decades. Dr Zhang has received numerous honours, including the Alfred P. Sloan Award, the U.S. National Science Foundation CAREER Award, and the University of Michigan Basic Science Research Award. Since 2015, he has been recognised eight times as a Thomson Reuters/Clarivate Analytics Highly Cited Researcher.
RESEARCH AREAS
Computational Biology
- Bioinformatics Algorithms
Artificial Intelligence
- Machine Learning
RESEARCH INTERESTS
AI and deep learning
Protein and RNA structure prediction
Protein and peptide design
AI-based drug discovery
RESEARCH PROJECTS

AI for Science
This research focuses on developing foundational artificial intelligence methods to advance scientific discovery. We study how modern machine learning models—such as deep learning, foundation models, and generative AI—can be tightly integrated with domain knowledge from physics, biology, and medicine to enable accurate modeling of biomolecular structures, interactions, and functions. This research direction is supported by national Foundational Research Capabilities (FRC) initiatives in Singapore, aiming to build long-term AI-driven scientific discovery platforms across disciplines.
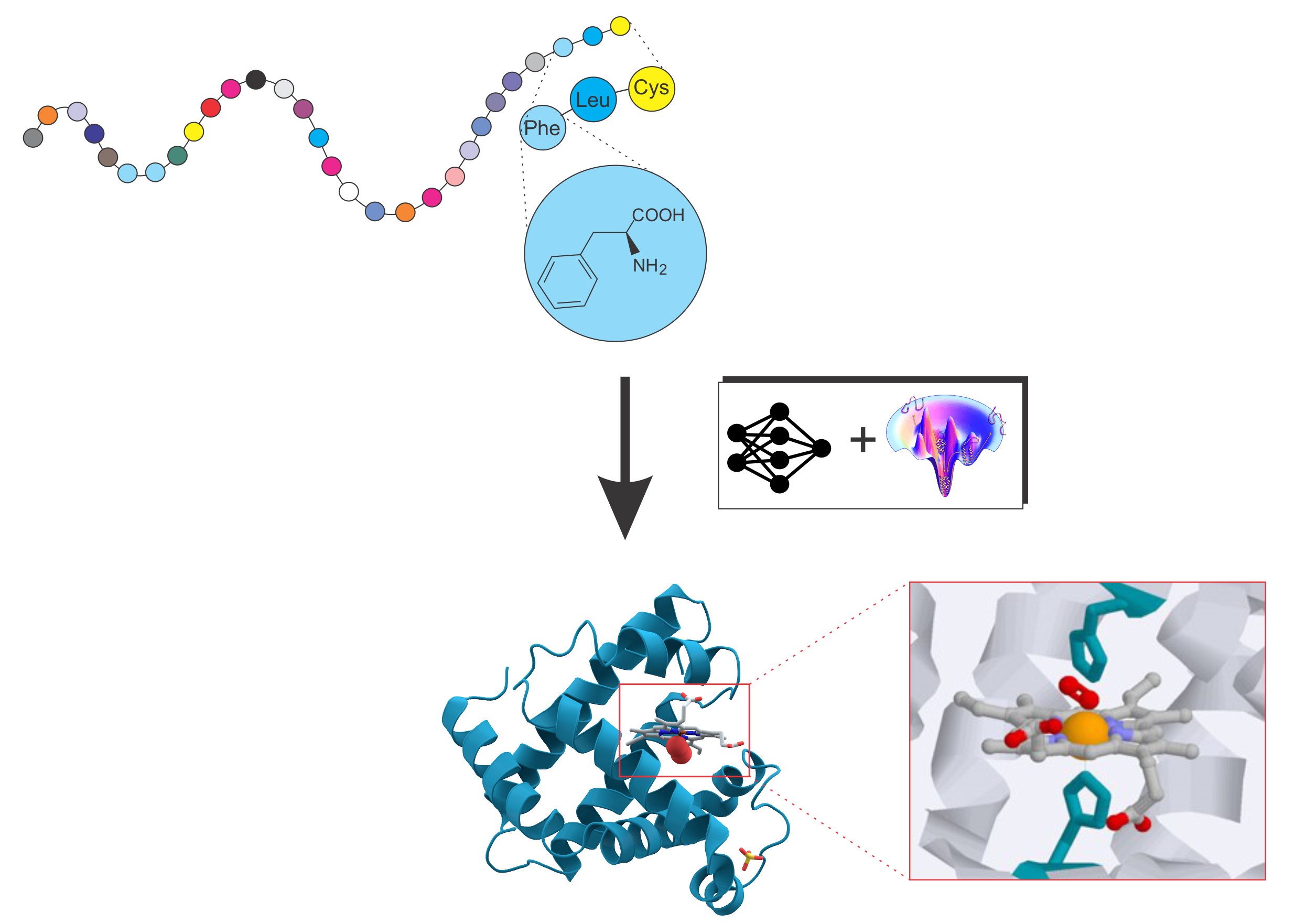
Protein folding and protein structure prediction
Deciphering the structure and function of proteins is central to modern biology and medicine. Our research develops pioneering computational frameworks that integrate advanced AI methodologies with physics-based force fields to accurately model protein structures and interactions. A central goal is to elucidate the fundamental relationships linking protein sequence, structure, dynamics, and function.
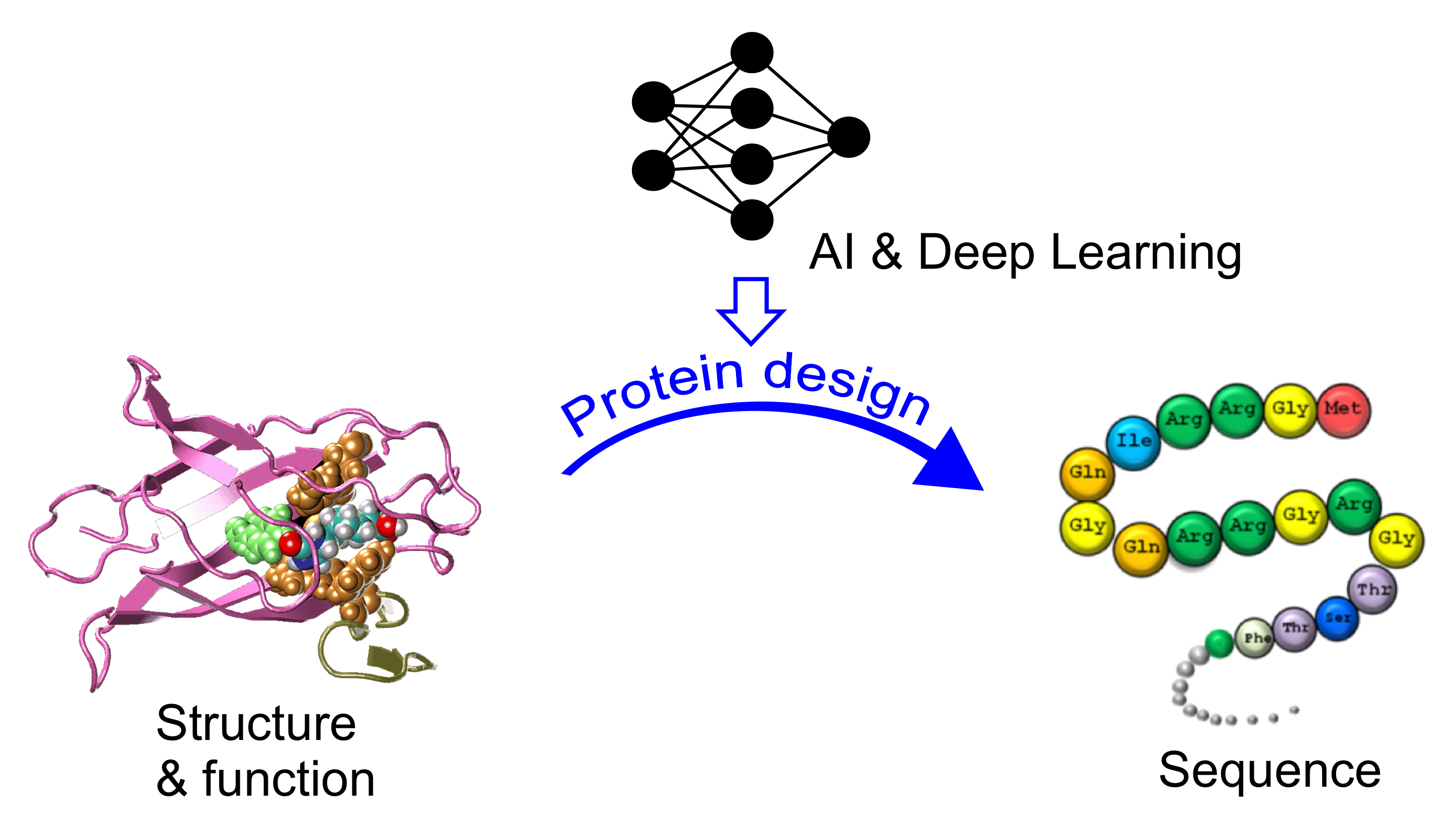
AI-based protein design and drug discovery
Natural proteins occupy a limited subset of the vast sequence–structure–function space shaped by evolution. This project develops AI- and deep learning–based frameworks for the rational design of novel proteins and peptides with tailored structures and functions. By integrating generative modeling with biophysical principles, we aim to expand the protein design space and enable new molecular scaffolds for biomedical and therapeutic applications.
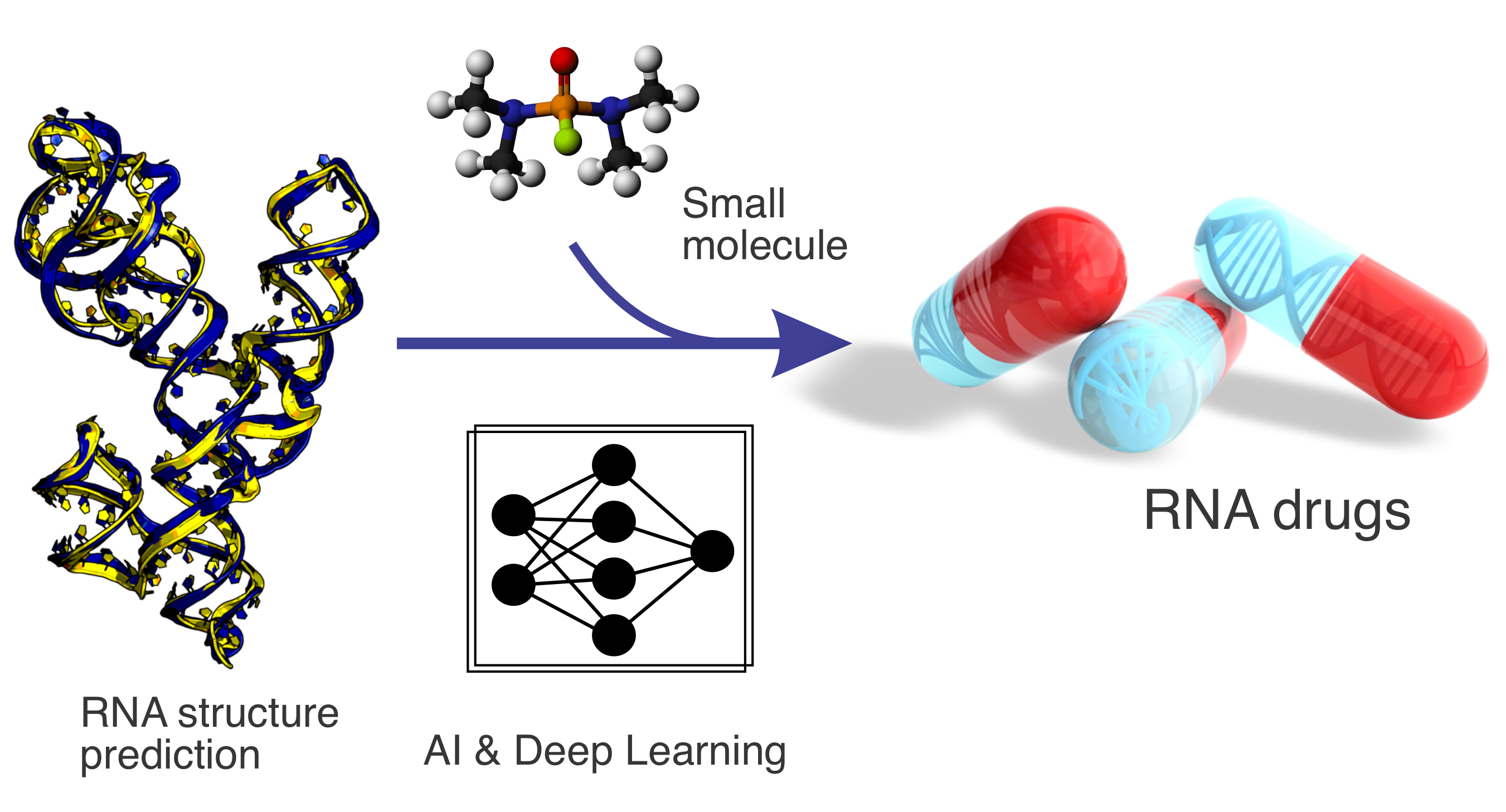
RNA structure prediction and small-molecule drug design
Non-coding RNAs play essential roles in regulating diverse cellular processes, yet their structural characterisation and ligand interactions remain challenging. This project develops advanced AI- and deep learning–based methods for RNA structure prediction and RNA–small-molecule interaction modeling. By integrating data-driven learning with biophysical principles, our work aims to enable RNA-targeted drug discovery and expand therapeutic opportunities beyond traditional protein-centric approaches.
RESEARCH GROUPS
TEACHING INNOVATIONS
SELECTED PUBLICATIONS
- (A full list of Dr. Yang Zhang's publications can be seen at https://scholar.google.com/citations?user=MtBs-kMAAAAJ)
- W Zheng, Q Wuyun, Y Li, Q Liu, X Zhou, C Peng, Y Zhu, L Freddolino, Y Zhang. Deep-learning-based single-domain and multidomain protein structure prediction with D-I-TASSER. Nature Biotechnology, https://doi.org/10.1038/s41587-025-02654-4 (2025).
- C Zhang, L Freddolino, Y Zhang. US-align: a graphic and command line protocol for quick and accurate comparisons of protein and nucleic acid structures. Nature Protocols, https://doi.org/10.1038/s41596-025-01189-x (2025).
- W Zheng, Q Wuyun, Y Li, C Zhang, PL Freddolino, Y Zhang. Improving deep learning protein monomer and complex structure prediction using DeepMSA2 with huge metagenomics data. Nature Methods, 21: 279-289 (2024).
- R Pearce, X Huang, GS Omenn, Y Zhang. De novo protein fold design through sequence-independent fragment assembly simulations. PNAS, 120: e2208275120 (2023).
- Y Li, C Zhang, C Feng, R Pearce, PL Freddolino, Y Zhang. Integrating end-to-end learning with deep geometrical potentials for ab initio RNA structure prediction. Nature Communications, 14: 5745 (2023).
- C Zhang, M Shine, AM Pyle, Y Zhang. US-align: Universal structure alignment of proteins, nucleic acids and macromolecular complexes. Nature Methods, 19: 1109-1115 (2022).
- X Zhou, W Zheng, Y Li, R Pearce, C Zhang, EW Bell, G Zhang, Y Zhang. I-TASSER-MTD: A deep-learning based platform for multi-domain protein structure and function prediction. Nature Protocols, 17: 2326-2353 (2022).
- X Zhou, Y Li, C Zhang, W Zheng, G Zhang, Y Zhang. Progressive assembly of multi-domain protein structures from cryo-EM density maps. Nature Computational Science, 2: 265-275 (2022).
- X Zhang, B Zhang, PL Freddolino, Y Zhang. CR-I-TASSER: Assemble protein structures from cryo-EM density maps using deep convolutional neural networks. Nature Methods, 19: 195-204 (2022).
- SM Mortuza, W Zheng, C Zhang, Y Li, R Pearce, Y Zhang. Improving fragment-based ab initio protein structure assembly using low-accuracy contact-map predictions. Nature Communications, 12: 5011 (2021).
- P Yang, W Zheng, K Ning, Y Zhang. Decoding the link of microbiome niches with homologous sequences enables accurately targeted protein structure prediction. PNAS, 118: e2110828118 (2021).
AWARDS & HONOURS
COURSES TAUGHT

