5 November 2020 – Several NUS Computing professors were recently awarded funding from the MOE Academic Research Funding (AcRF) Tier 2 scheme. Among the grant recipients were Professor Dong Jin Song, Professor Wing-Kin Sung and Assistant Professor Lee Gim Hee, all from the Department of Computer Science.
Through the funding, MOE aims to support and advance research conducted in autonomous universities in Singapore through the creation of new knowledge, or knowledge that “challenges current understanding”.
The MOE AcRF Tier 2 scheme provides researchers with funding between SGD$500,000 and SGD$1 million.
Improving software debugging
Professor Dong Jin Song is developing an execution-trace based technique and platform to help programmers automatically locate and explain the root cause of regression bugs.
Software regression bugs may cause a previously operating feature in the software to stop working after a part of the code is modified. This results in the new version of the modified software failing the test case, a software testing procedure.
Locating regression bugs may be challenging when there are a large number of modifications made between the previous and current versions of a software, Professor Dong explained. Manual debugging usually requires repetitive comparisons and analysis between the two versions, which is time consuming and tedious.
Current and traditional approaches typically leverage on several program analysis techniques which may inaccurately point out these failure-inducing changes. An inaccurate report of failure-inducing changes may be due in part to human error, when programmers forget to apply the changes made to the software code. Even if failure-inducing changes are reported accurately, the programmers may still need to trawl through the code and figure out a solution to fix the bug.
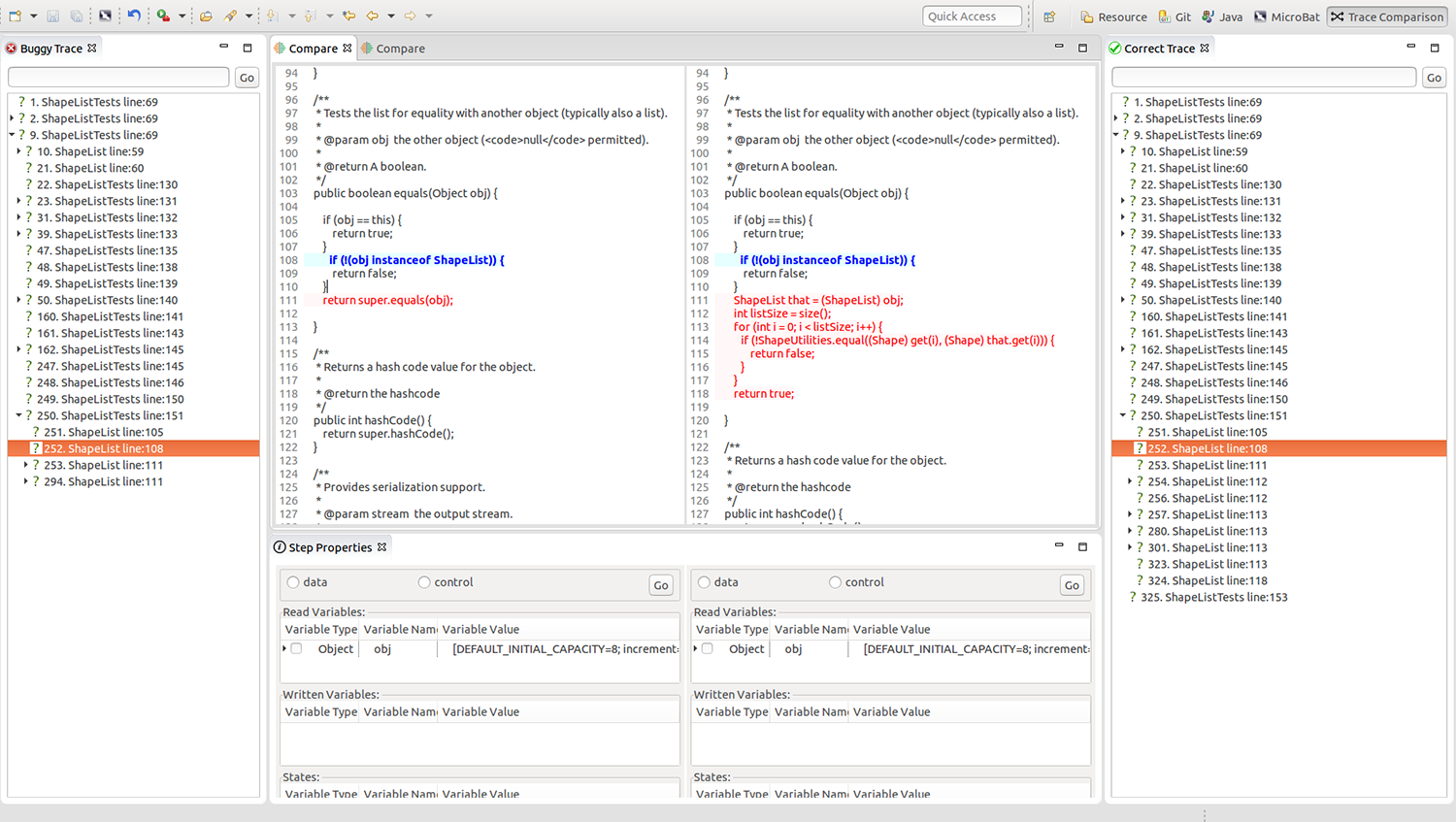
Professor Dong’s project aims to develop a user-friendly platform that utilises an execution-trace based technique, “a variant of time-travelling debugging”, which collects and records information about what is happening in a program. Programmers may conduct an analysis using this information provided via an interactive user interface.
Professor Dong is working with Dr Lin Yun, a Senior Research Fellow at NUS Computing, on this ongoing research project.
“We believe that this tool will benefit the programmers in IT industries and mitigate the debugging challenges in their daily tasks,” Prof Dong said. “The support of this grant will definitely speed up our progress on software debugging research.”
Discovering gene variations in an Asian population
Professor Sung is working on methods to study genomic variants in a sub-population, such as the Asian or Singapore population. Genomic variations refer to the differences between genomes (a living being’s complete set of DNA, including its genes), which may have an impact on one’s health.
Current methods for studying genomic variations typically use the standard reference genome, which is different from an Asian or Singaporean genome.
“We observed that the Asian (or Singaporean) genome is not similar to the standard reference genome. By using the current reference genome, we may miss out on discovering a lot of genomic variants and identify false variants instead,” said Professor Sung.

With this research project, they aim to develop tools that will increase the accuracy of identifying genomic variations. Better understanding of the Asian genome will enable researchers to locate the causes behind certain diseases that are present in certain races, and provide better medical solutions and healthcare for patients.
“The information of our genome can tell us a lot of things. For example, they can tell us why Asians are more likely to have liver cancer, or why some people are resistant to some drugs. With this funding, we hope that we can contribute to improve the understanding of our human genome,” Professor Sung said. “The funding awarded is a great motivation to our team.”
Lowering barriers to deep learning for 3D computer vision
To lower the barrier of entry for deep learning on 3D computer vision-related tasks such as 3D object detection and 3D semantic segmentation, Assistant Professor Lee Gim Hee wants to advance the research on alternative learning strategies for 3D computer vision.
“The objective of 3D computer vision is to gain geometric and semantic understanding of the 3D world from sensory data such as camera images, and/or 3D point clouds from Lidar scans,” A/P Lee explained. “3D computer vision is very useful for various industries, such as facilitating autonomous or assisted driving in automobiles, improving wearable devices used in healthcare and education, or metrological devices in building and construction.”
Deep learning is an artificial intelligence function that mimics the biological structure and functioning of a human brain. A deep learning network consists of many layers of nodes interconnected by learnable parameters that are trained by being shown large amounts of examples, which are carefully annotated ground truth data. This process is known as strongly supervised learning, and it has been successfully used to boost the performance of many 3D computer vision tasks in the recent years.
However, such annotated 3D datasets are difficult to obtain and expensive in large quantities, as manpower is required to manually label them, explained A/P Lee. The challenge is further aggravated by the amodality of most 3D data, where only part of the scene or object is available (e.g. a cat hiding behind a fence). In contrast, humans do not require strong supervision from large amounts of annotated data to learn. Besides learning from being shown examples, humans learn to identify new instances of the same object from geometrical cues, such as shape and size. For instance, a toddler is able to easily identify a cup after seeing one or two examples of different cups.
Together with his team, AP Lee aims to alleviate the need for large amounts of strongly-labelled datasets when it comes to deep learning for 3D computer vision. He is tapping on 3D geometric information that inherently exists in any 3D scene or object, and aims to fuse this information as priors into alternative learning strategies such as semi-supervised, weakly-supervised, self-supervised, or few-shot learning to decrease overreliance on large amounts of labelled data.
“We believe that a successful outcome of this research will lower the barrier of using deep learning effectively on 3D computer vision-related tasks in the industry,” said A/P Lee. “I am thankful for the funding that allows us to continue our research. This means MOE recognises the project’s potential impact on Singapore’s development as a smart nation, and in advancing the fields of computer vision and robotics.”