To say that the human body is an intricate complicated system would be an understatement. When one thing goes wrong, others often follow suit. So in 1970 when Alvan Feinstein first coined the term ‘comorbidity’ — to refer to a person having multiple diseases at the same time — it wasn’t too revolutionary a concept.
Back then, the renowned American physician was trying to describe how patients with rheumatic fever also seemed to suffer from other diseases. Today, comorbidities abound: diabetes is linked with a higher risk of heart disease and renal failure, anxiety frequently occurs with mood disorders, and the list goes on.
“Many patients, especially the ones who have chronic diseases, tend to have two or more problems at the same time,” says Vaibhav Rajan, an assistant professor at NUS Computing who researches healthcare informatics. The earlier that doctors can predict a patient is likely to develop other diseases, the better for all parties involved — patients can proactively take steps to lower their risk, doctors can prescribe preventive treatments, and the overall clinical and economic burden of a disease is reduced.
But it isn’t always easy for physicians to diagnose a disease, much less identify all the possible conditions that could develop as a result of that disease, or ones that can co-exist simultaneously with it. And when the disease is rare, things are even harder.
The reality is that clinical knowledge is increasing at a rapid rate — more than 800,000 new journal articles are added to the biomedical database MEDLINE every year. “Nobody can read the millions of articles that are there,” he says. “Doctors may not have the time to read and be up to date on all the findings reported.”
So in recent years, researchers have devised various diagnostic decision support tools to help with such predictions. Multi-Disease Predictive Analytics (MDPA) models use historical data from electronic health records of real-life patients to model the correlations between different diseases and predict their risks simultaneously. Through this analysis, the models can predict the likelihood a patient with a disease X will develop diseases Y and Z, for instance.
MDPA models can be developed using machine learning techniques. As with all such methods, the more data there is, the more accurate the predictions. “For very common diseases, there’s typically a lot of data in hospitals so you can easily build a model and get predictions,” says Rajan. “But for rare diseases, it’s difficult since there are much fewer patients from past records whose data can be utilised in model building.”
Looking to a different source
In 2018, Rajan — together with his NUS Computing colleague Shaw Senior Professor Bernard Tan and students Lin Qiu and Sruthi Gorantla — began working to overcome this problem.
For Rajan, it was a challenge right up his alley. “The general theme of research in my lab is to develop methods to integrate different sources of information to build predictive models to aid various clinical decisions.”
MDPA models make use of patients’ medical histories, but they do not sufficiently use another data source that is rich with information: knowledge of disease correlations that have been recorded in biomedical literature. Could this additional data be automatically used by the models, Rajan and his team wondered. If so, it would help improve their predictive accuracy, especially for rare diseases.
“Biomedical literature is the primary source of clinical knowledge but comes in an unstructured form — as journal articles — and it’s difficult for machine learning models to use them directly,” he explains. However, there is ongoing research to use AI techniques to ‘understand’ the literature and represent them in the form of knowledge graphs.
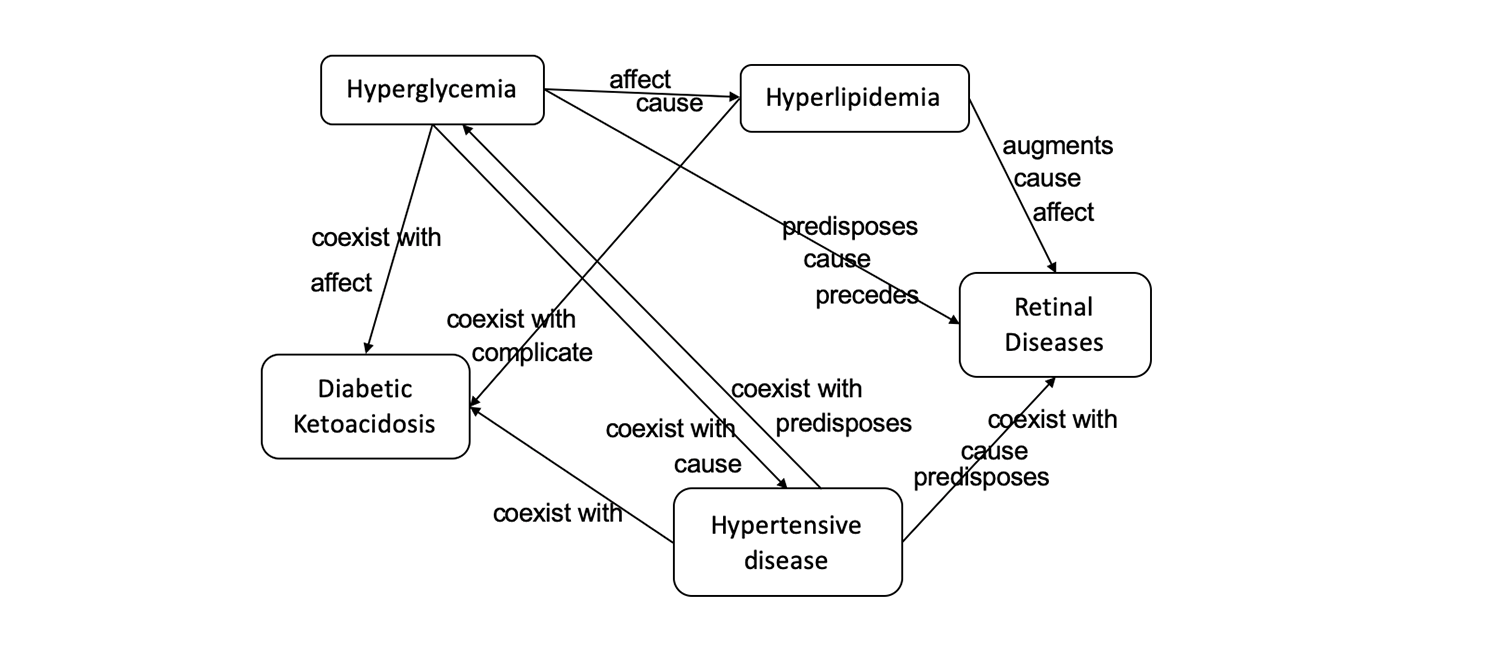
Think of each knowledge graph as a mind map of sorts, comprising numerous nodes connected by pathways. Each node is a clinical concept, such as disease or a drug. Pathways connecting nodes indicate relationships between two factors, for example whether a condition is considered to be a risk factor for the disease or if a certain drug can be used to treat it.
“What we tried to do was to develop a method that could take advantage of these graphs and see how we could combine it with MDPA models,” says Rajan. Most previous methods that have tried to integrate knowledge graphs with MDPA models have done so when the model is being trained. The novelty of Rajan’s approach is the use of the knowledge graphs during the prediction phase to refine a model’s outputs.
“This decoupling allows our approach to be used with any kind of MDPA model,” he says. “As biomedical literature grows, our approach can be updated periodically and applied on the MDPA model without re-training the model.”
An added benefit: the new method makes use of the entire biomedical knowledge graph, which means it is more general than existing techniques that have been designed to employ specific subsets of the graphs.

Boosting prediction power
Rajan and the team called their new method the Knowledge-Aware Approach (KAA). To test how well KAA worked, they applied it to a real-life dataset of more than 7,500 people, taken from a publicly available dataset of intensive care patients in Boston. The patients were chosen because they were admitted more than once to the same hospital, and had two coexisting conditions that were one of the 20 most common diseases or the 10 rarest ones.
Rajan’s analysis, described in a paper published this year, demonstrates how when KAA is applied to commonly used predictive models, it improves a doctor’s ability to recognise correlations between two diseases and to prescribe preventive treatments appropriately. This was the case for both common and rare diseases.
Since then, Rajan has continued to explore the use of knowledge graphs for use in other models. In 2020, he published a paper examining how they can be used to predict adverse drug events. He is now looking to see how to improve the way in which information is extracted from these graphs.
“We’re getting more and more data from biomedical literature, and the techniques used to extract this knowledge are also improving,” says Rajan. “Which means that our models should also improve in terms of predictive accuracy and interpretability.”
“Once these methods and systems improve, and are well integrated into clinical workflows, I think that diagnosis could be increasingly more AI-driven in the future,” he says.
[1] Kilicoglu, Halil, et al. “SemMedDB: a PubMed-scale repository of biomedical semantic predications.” Bioinformatics 28.23 (2012): 3158-3160.
Paper: Multi-disease Predictive Analytics: A Clinical Knowledge-aware Approach